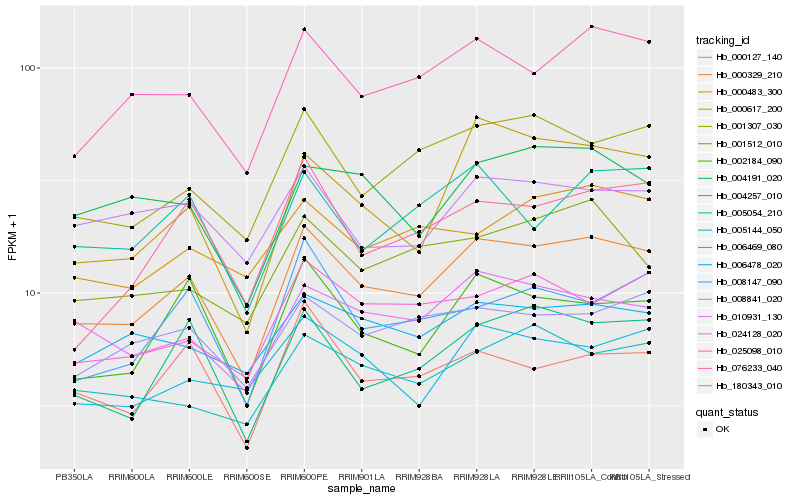
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_210 |
0.0 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 2 |
Hb_076233_040 |
0.0765234345 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001307_030 |
0.0831767969 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 4 |
Hb_004257_010 |
0.0855432758 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 5 |
Hb_002184_090 |
0.0875837924 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_024128_020 |
0.0883507354 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000617_200 |
0.0939301768 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 8 |
Hb_008841_020 |
0.0939448266 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631075 isoform X1 [Jatropha curcas] |
| 9 |
Hb_025098_010 |
0.0957218846 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 10 |
Hb_000127_140 |
0.0962481861 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 11 |
Hb_005054_210 |
0.0968993616 |
- |
- |
hypothetical protein PRUPE_ppa026456mg [Prunus persica] |
| 12 |
Hb_004191_020 |
0.0970147088 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 13 |
Hb_005144_050 |
0.0971035924 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 isoform X2 [Vitis vinifera] |
| 14 |
Hb_006478_020 |
0.0971167427 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 15 |
Hb_001512_010 |
0.0995014328 |
- |
- |
PREDICTED: uncharacterized protein LOC105633759 [Jatropha curcas] |
| 16 |
Hb_180343_010 |
0.1006924819 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 17 |
Hb_006469_080 |
0.101159837 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008147_090 |
0.1015547843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000483_300 |
0.1027689435 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 20 |
Hb_010931_130 |
0.1028048061 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |