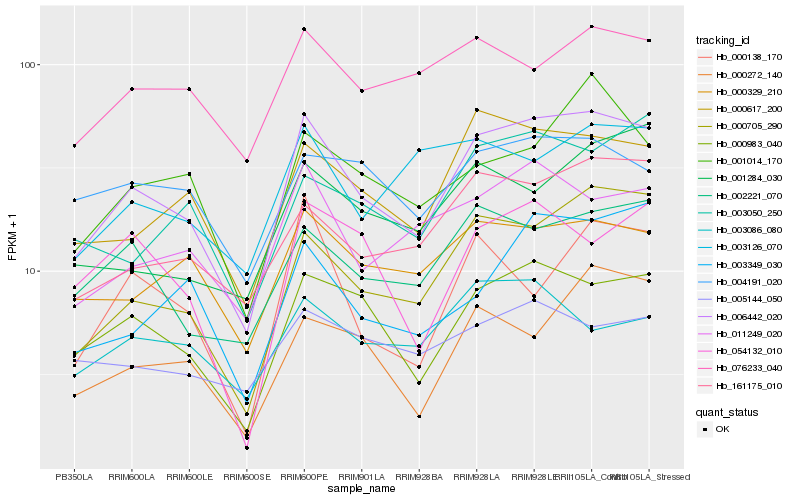
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006442_020 |
0.0 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 2 |
Hb_000983_040 |
0.0973371512 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 3 |
Hb_000705_290 |
0.0976610633 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |
| 4 |
Hb_000617_200 |
0.1125622411 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 5 |
Hb_161175_010 |
0.1199366436 |
- |
- |
PREDICTED: vesicle-associated protein 1-1 [Jatropha curcas] |
| 6 |
Hb_002221_070 |
0.1238372586 |
- |
- |
beta-glucanase, putative [Ricinus communis] |
| 7 |
Hb_011249_020 |
0.1243223468 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 8 |
Hb_000329_210 |
0.1279452522 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 9 |
Hb_001014_170 |
0.1358661081 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000272_140 |
0.1386247453 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 11 |
Hb_003050_250 |
0.1403500168 |
- |
- |
PREDICTED: uncharacterized protein At2g34460, chloroplastic [Jatropha curcas] |
| 12 |
Hb_076233_040 |
0.1416713783 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001284_030 |
0.1434113764 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_054132_010 |
0.1443058588 |
- |
- |
PREDICTED: transmembrane protein 136-like isoform X2 [Pyrus x bretschneideri] |
| 15 |
Hb_003126_070 |
0.1449167185 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003349_030 |
0.1452309762 |
- |
- |
PREDICTED: uncharacterized protein LOC105628283 [Jatropha curcas] |
| 17 |
Hb_004191_020 |
0.1475799438 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 18 |
Hb_000138_170 |
0.1497434395 |
- |
- |
putative plastid-lipid-associated protein, partial [Hevea brasiliensis] |
| 19 |
Hb_003086_080 |
0.150721837 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 20 |
Hb_005144_050 |
0.1520317889 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 isoform X2 [Vitis vinifera] |