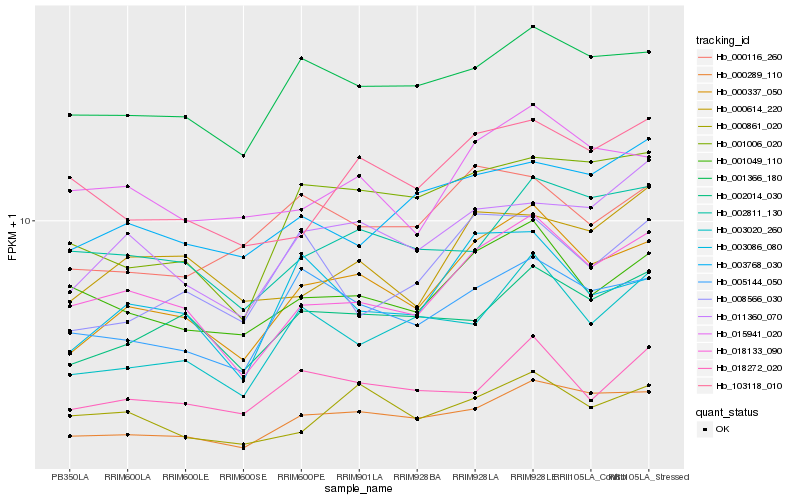
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000337_050 |
0.0 |
- |
- |
PREDICTED: chloroplast envelope membrane protein [Jatropha curcas] |
| 2 |
Hb_000289_110 |
0.077748374 |
- |
- |
PREDICTED: cysteine synthase-like [Musa acuminata subsp. malaccensis] |
| 3 |
Hb_018133_090 |
0.0809949081 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003086_080 |
0.1001968809 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 5 |
Hb_001366_180 |
0.1008366486 |
- |
- |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_005144_050 |
0.1017264471 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 isoform X2 [Vitis vinifera] |
| 7 |
Hb_003020_260 |
0.1036359233 |
- |
- |
PREDICTED: metal tolerance protein B [Jatropha curcas] |
| 8 |
Hb_002014_030 |
0.1040606916 |
- |
- |
hypothetical protein JCGZ_07251 [Jatropha curcas] |
| 9 |
Hb_001049_110 |
0.1050690119 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_011360_070 |
0.10671165 |
- |
- |
PREDICTED: zinc finger RNA-binding protein [Jatropha curcas] |
| 11 |
Hb_001006_020 |
0.1067728187 |
- |
- |
PREDICTED: WW domain-binding protein 4 [Jatropha curcas] |
| 12 |
Hb_015941_020 |
0.1070436962 |
- |
- |
- |
| 13 |
Hb_000861_020 |
0.1170434992 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 14 |
Hb_000116_260 |
0.117223619 |
- |
- |
PREDICTED: uncharacterized protein LOC105628572 [Jatropha curcas] |
| 15 |
Hb_018272_020 |
0.1179691714 |
- |
- |
PREDICTED: small glutamine-rich tetratricopeptide repeat-containing protein alpha [Jatropha curcas] |
| 16 |
Hb_103118_010 |
0.1185732085 |
- |
- |
PREDICTED: uncharacterized protein LOC105643231 [Jatropha curcas] |
| 17 |
Hb_008566_030 |
0.1187478949 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 18 |
Hb_003768_030 |
0.11897874 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_000614_220 |
0.118993266 |
- |
- |
PREDICTED: uncharacterized protein LOC105644756 [Jatropha curcas] |
| 20 |
Hb_002811_130 |
0.1190321397 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |