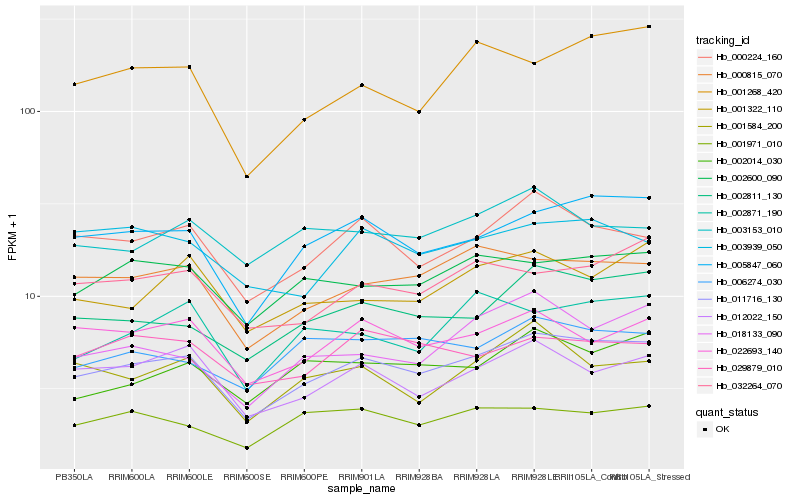
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011716_130 |
0.0 |
- |
- |
PREDICTED: probable acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, mitochondrial isoform X5 [Populus euphratica] |
| 2 |
Hb_005847_060 |
0.0607879861 |
- |
- |
hypothetical protein POPTR_0006s02920g [Populus trichocarpa] |
| 3 |
Hb_002811_130 |
0.0750152745 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000815_070 |
0.0765692515 |
- |
- |
PREDICTED: methyltransferase-like protein 7A [Jatropha curcas] |
| 5 |
Hb_000224_160 |
0.0770046278 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_002600_090 |
0.080415886 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_012022_150 |
0.0843850814 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |
| 8 |
Hb_022693_140 |
0.0864002857 |
- |
- |
PREDICTED: uncharacterized protein LOC105632654 isoform X1 [Jatropha curcas] |
| 9 |
Hb_032264_070 |
0.0874019399 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001322_110 |
0.0916400219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001971_010 |
0.0923852479 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 12 |
Hb_006274_030 |
0.0929874147 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 13 |
Hb_018133_090 |
0.0930596142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002014_030 |
0.0931900024 |
- |
- |
hypothetical protein JCGZ_07251 [Jatropha curcas] |
| 15 |
Hb_001268_420 |
0.094313561 |
- |
- |
PREDICTED: uncharacterized protein LOC105630158 [Jatropha curcas] |
| 16 |
Hb_002871_190 |
0.095393152 |
- |
- |
PREDICTED: uncharacterized protein LOC105628867 [Jatropha curcas] |
| 17 |
Hb_003939_050 |
0.0971115729 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_001584_200 |
0.0973085673 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 19 |
Hb_029879_010 |
0.0978137424 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |
| 20 |
Hb_003153_010 |
0.0982642075 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |