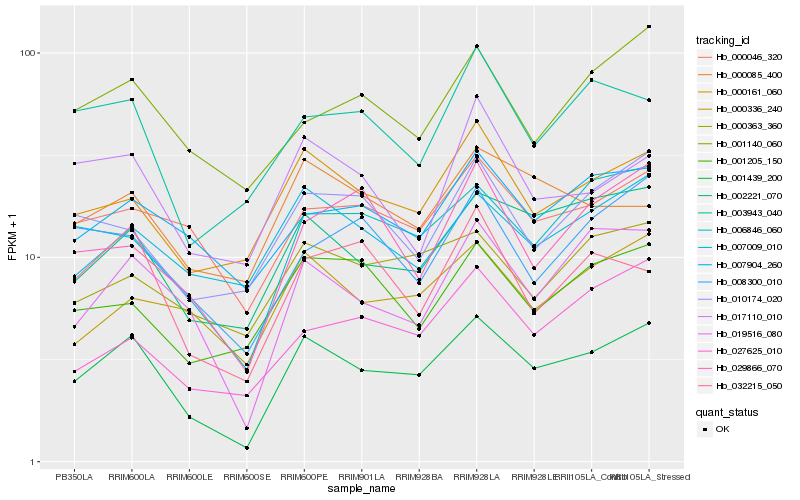
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001439_200 |
0.0 |
- |
- |
Transmembrane protein Tmp21 precursor, putative [Ricinus communis] |
| 2 |
Hb_002221_070 |
0.1128660862 |
- |
- |
beta-glucanase, putative [Ricinus communis] |
| 3 |
Hb_019516_080 |
0.1145929773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000161_060 |
0.1221170757 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 [Jatropha curcas] |
| 5 |
Hb_006846_060 |
0.1247876095 |
- |
- |
inositol hexaphosphate kinase, putative [Ricinus communis] |
| 6 |
Hb_000046_320 |
0.1349304218 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 7 |
Hb_007904_260 |
0.1385275155 |
- |
- |
PREDICTED: uncharacterized protein LOC105644715 [Jatropha curcas] |
| 8 |
Hb_000336_240 |
0.1385360898 |
transcription factor |
TF Family: mTERF |
LOC100285792 [Zea mays] |
| 9 |
Hb_007009_010 |
0.1397299081 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675 [Populus euphratica] |
| 10 |
Hb_032215_050 |
0.1426005135 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 11 |
Hb_000363_360 |
0.1435580547 |
- |
- |
hypothetical protein PRUPE_ppa009146mg [Prunus persica] |
| 12 |
Hb_010174_020 |
0.1437732934 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_003943_040 |
0.1464025738 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_008300_010 |
0.1473251675 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG13 homolog [Jatropha curcas] |
| 15 |
Hb_001140_060 |
0.1473477684 |
- |
- |
PREDICTED: ADP-ribosylation factor-related protein 1 [Prunus mume] |
| 16 |
Hb_001205_150 |
0.1477075573 |
- |
- |
Uncharacterized protein TCM_000403 [Theobroma cacao] |
| 17 |
Hb_017110_010 |
0.1482434001 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g55760 [Jatropha curcas] |
| 18 |
Hb_027625_010 |
0.1489487061 |
- |
- |
PREDICTED: protein sym-1-like [Jatropha curcas] |
| 19 |
Hb_000085_400 |
0.1490116195 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 20 |
Hb_029866_070 |
0.1495392602 |
- |
- |
PREDICTED: myosin IB heavy chain [Jatropha curcas] |