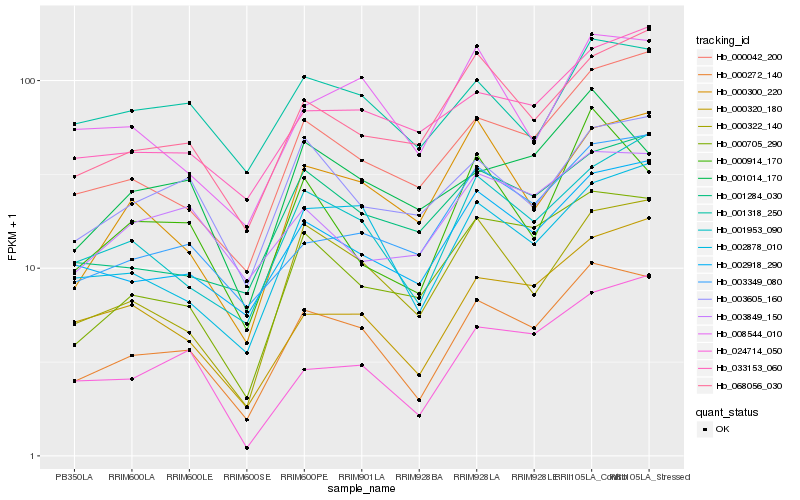
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000272_140 |
0.0 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 2 |
Hb_002878_010 |
0.1028390605 |
- |
- |
- |
| 3 |
Hb_002918_290 |
0.1053485312 |
- |
- |
PREDICTED: uncharacterized protein LOC105649583 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000705_290 |
0.1057224755 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |
| 5 |
Hb_001953_090 |
0.1073874547 |
- |
- |
PREDICTED: membrane-anchored ubiquitin-fold protein 3 [Jatropha curcas] |
| 6 |
Hb_000042_200 |
0.1105245861 |
- |
- |
PREDICTED: uncharacterized protein LOC100252136 isoform X1 [Vitis vinifera] |
| 7 |
Hb_000322_140 |
0.11394725 |
- |
- |
hypothetical protein RCOM_1047200 [Ricinus communis] |
| 8 |
Hb_068056_030 |
0.1216663646 |
- |
- |
ATP synthase epsilon chain, mitochondrial, putative [Ricinus communis] |
| 9 |
Hb_000320_180 |
0.1243361599 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 10 |
Hb_003349_080 |
0.1265211759 |
- |
- |
PREDICTED: uncharacterized protein LOC105628287 [Jatropha curcas] |
| 11 |
Hb_003605_160 |
0.1271265451 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 12 |
Hb_001284_030 |
0.1296141456 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_008544_010 |
0.1309936373 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000300_220 |
0.1312505605 |
- |
- |
PREDICTED: uncharacterized protein LOC105632569 [Jatropha curcas] |
| 15 |
Hb_033153_060 |
0.1316203183 |
- |
- |
Transcription elongation factor, putative [Ricinus communis] |
| 16 |
Hb_001014_170 |
0.1328586755 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000914_170 |
0.1334818502 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 18 |
Hb_024714_050 |
0.1340487712 |
- |
- |
hypothetical protein CICLE_v100188992mg, partial [Citrus clementina] |
| 19 |
Hb_003849_150 |
0.1343972594 |
- |
- |
PREDICTED: uncharacterized protein LOC105644698 [Jatropha curcas] |
| 20 |
Hb_001318_250 |
0.1345282801 |
- |
- |
PREDICTED: uncharacterized protein LOC105642666 [Jatropha curcas] |