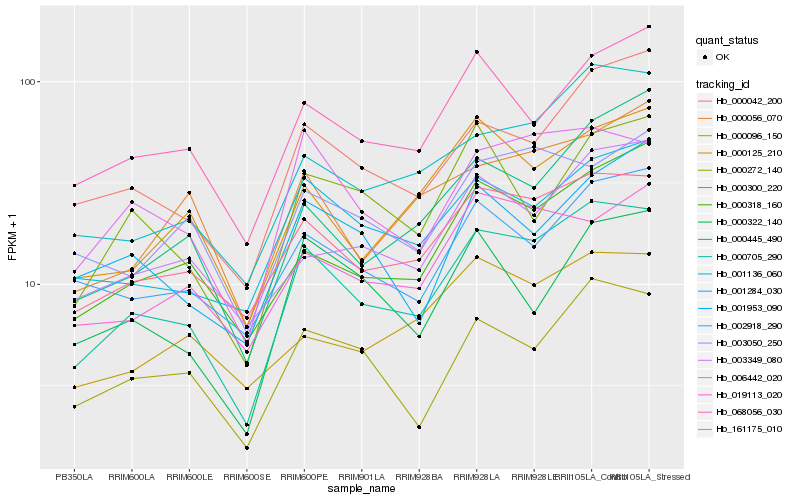
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000705_290 |
0.0 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |
| 2 |
Hb_161175_010 |
0.0861583398 |
- |
- |
PREDICTED: vesicle-associated protein 1-1 [Jatropha curcas] |
| 3 |
Hb_006442_020 |
0.0976610633 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 4 |
Hb_001136_060 |
0.1052895692 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 5 |
Hb_000272_140 |
0.1057224755 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 6 |
Hb_000042_200 |
0.1060793804 |
- |
- |
PREDICTED: uncharacterized protein LOC100252136 isoform X1 [Vitis vinifera] |
| 7 |
Hb_001284_030 |
0.1063496509 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000318_160 |
0.1127300387 |
- |
- |
PREDICTED: uncharacterized protein LOC105634239 [Jatropha curcas] |
| 9 |
Hb_068056_030 |
0.1132840024 |
- |
- |
ATP synthase epsilon chain, mitochondrial, putative [Ricinus communis] |
| 10 |
Hb_002918_290 |
0.1172590384 |
- |
- |
PREDICTED: uncharacterized protein LOC105649583 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000096_150 |
0.1223551217 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_003349_080 |
0.1240945958 |
- |
- |
PREDICTED: uncharacterized protein LOC105628287 [Jatropha curcas] |
| 13 |
Hb_019113_020 |
0.12438791 |
- |
- |
hypothetical protein JCGZ_13398 [Jatropha curcas] |
| 14 |
Hb_000300_220 |
0.1267451995 |
- |
- |
PREDICTED: uncharacterized protein LOC105632569 [Jatropha curcas] |
| 15 |
Hb_003050_250 |
0.1278951789 |
- |
- |
PREDICTED: uncharacterized protein At2g34460, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000056_070 |
0.1293306731 |
- |
- |
- |
| 17 |
Hb_000322_140 |
0.1294993049 |
- |
- |
hypothetical protein RCOM_1047200 [Ricinus communis] |
| 18 |
Hb_000125_210 |
0.1298927981 |
- |
- |
PREDICTED: electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000445_490 |
0.132713371 |
- |
- |
- |
| 20 |
Hb_001953_090 |
0.1331584734 |
- |
- |
PREDICTED: membrane-anchored ubiquitin-fold protein 3 [Jatropha curcas] |