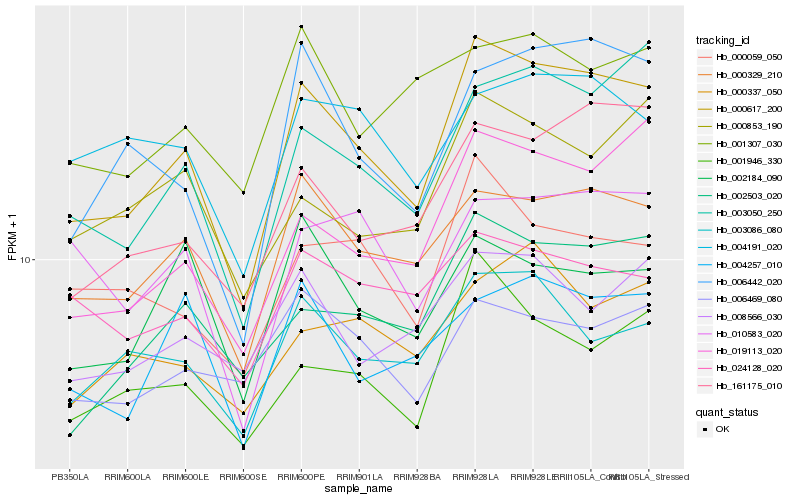
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000617_200 |
0.0 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 2 |
Hb_000329_210 |
0.0939301768 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 3 |
Hb_003050_250 |
0.1057894349 |
- |
- |
PREDICTED: uncharacterized protein At2g34460, chloroplastic [Jatropha curcas] |
| 4 |
Hb_019113_020 |
0.1064796996 |
- |
- |
hypothetical protein JCGZ_13398 [Jatropha curcas] |
| 5 |
Hb_161175_010 |
0.1094661542 |
- |
- |
PREDICTED: vesicle-associated protein 1-1 [Jatropha curcas] |
| 6 |
Hb_002503_020 |
0.1101860773 |
- |
- |
Phosphatase 2C family protein, putative [Theobroma cacao] |
| 7 |
Hb_006442_020 |
0.1125622411 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 8 |
Hb_003086_080 |
0.1134658097 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 9 |
Hb_001946_330 |
0.1158423677 |
- |
- |
PREDICTED: transmembrane protein 128 [Jatropha curcas] |
| 10 |
Hb_000059_050 |
0.1202729945 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 11 |
Hb_004257_010 |
0.1235840221 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 12 |
Hb_008566_030 |
0.1246089664 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 13 |
Hb_000853_190 |
0.1274578106 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 14 |
Hb_006469_080 |
0.1287781413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000337_050 |
0.1297044839 |
- |
- |
PREDICTED: chloroplast envelope membrane protein [Jatropha curcas] |
| 16 |
Hb_010583_020 |
0.1302083365 |
- |
- |
PREDICTED: uncharacterized protein LOC105646948 [Jatropha curcas] |
| 17 |
Hb_002184_090 |
0.1307064309 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_004191_020 |
0.1316267674 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 19 |
Hb_001307_030 |
0.1319093291 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 20 |
Hb_024128_020 |
0.1329662183 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |