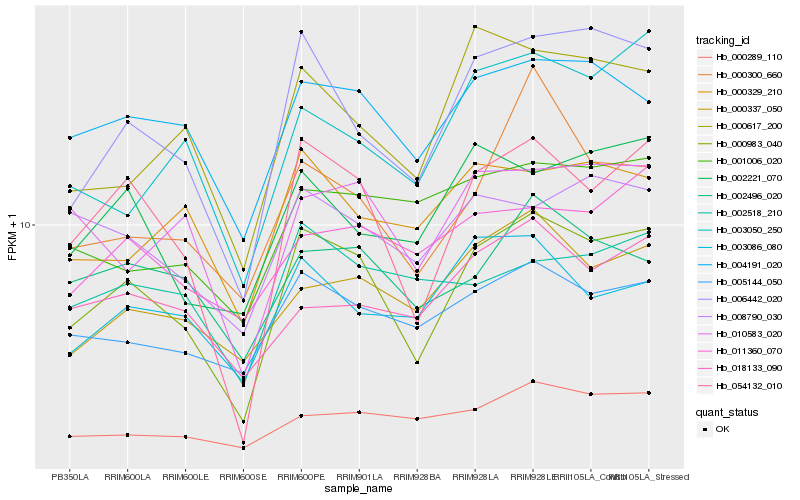
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000983_040 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 2 |
Hb_054132_010 |
0.0739294052 |
- |
- |
PREDICTED: transmembrane protein 136-like isoform X2 [Pyrus x bretschneideri] |
| 3 |
Hb_006442_020 |
0.0973371512 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 4 |
Hb_004191_020 |
0.1149010864 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 5 |
Hb_005144_050 |
0.1270300683 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 isoform X2 [Vitis vinifera] |
| 6 |
Hb_000337_050 |
0.1334073053 |
- |
- |
PREDICTED: chloroplast envelope membrane protein [Jatropha curcas] |
| 7 |
Hb_000617_200 |
0.1334673556 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 8 |
Hb_010583_020 |
0.1336429943 |
- |
- |
PREDICTED: uncharacterized protein LOC105646948 [Jatropha curcas] |
| 9 |
Hb_002221_070 |
0.134152709 |
- |
- |
beta-glucanase, putative [Ricinus communis] |
| 10 |
Hb_008790_030 |
0.1375867174 |
- |
- |
PREDICTED: probable magnesium transporter NIPA9 [Jatropha curcas] |
| 11 |
Hb_002496_020 |
0.1381810405 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_003086_080 |
0.1416501239 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 13 |
Hb_000329_210 |
0.1419069811 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 14 |
Hb_003050_250 |
0.1425259612 |
- |
- |
PREDICTED: uncharacterized protein At2g34460, chloroplastic [Jatropha curcas] |
| 15 |
Hb_011360_070 |
0.1430570712 |
- |
- |
PREDICTED: zinc finger RNA-binding protein [Jatropha curcas] |
| 16 |
Hb_001006_020 |
0.1453751488 |
- |
- |
PREDICTED: WW domain-binding protein 4 [Jatropha curcas] |
| 17 |
Hb_018133_090 |
0.1464519168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000300_660 |
0.1465326686 |
- |
- |
unknown [Glycine max] |
| 19 |
Hb_000289_110 |
0.1468905718 |
- |
- |
PREDICTED: cysteine synthase-like [Musa acuminata subsp. malaccensis] |
| 20 |
Hb_002518_210 |
0.1471713734 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 2 [Jatropha curcas] |