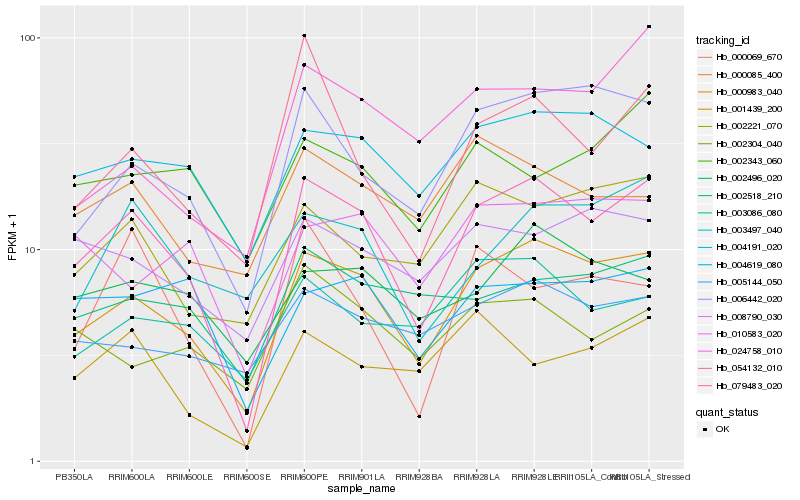
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054132_010 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 136-like isoform X2 [Pyrus x bretschneideri] |
| 2 |
Hb_000983_040 |
0.0739294052 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 3 |
Hb_006442_020 |
0.1443058588 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 4 |
Hb_004191_020 |
0.1612556621 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 5 |
Hb_079483_020 |
0.1614295625 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_004619_080 |
0.1651780908 |
- |
- |
hypothetical protein PRUPE_ppa011369mg [Prunus persica] |
| 7 |
Hb_002221_070 |
0.1684894295 |
- |
- |
beta-glucanase, putative [Ricinus communis] |
| 8 |
Hb_003497_040 |
0.169030496 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_001439_200 |
0.1714399399 |
- |
- |
Transmembrane protein Tmp21 precursor, putative [Ricinus communis] |
| 10 |
Hb_002518_210 |
0.1715580035 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 2 [Jatropha curcas] |
| 11 |
Hb_010583_020 |
0.1717219651 |
- |
- |
PREDICTED: uncharacterized protein LOC105646948 [Jatropha curcas] |
| 12 |
Hb_008790_030 |
0.1726363859 |
- |
- |
PREDICTED: probable magnesium transporter NIPA9 [Jatropha curcas] |
| 13 |
Hb_005144_050 |
0.1737844802 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 isoform X2 [Vitis vinifera] |
| 14 |
Hb_002496_020 |
0.1751533031 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_003086_080 |
0.1754098403 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000069_670 |
0.1756704809 |
- |
- |
PREDICTED: uncharacterized protein LOC105642887 [Jatropha curcas] |
| 17 |
Hb_002343_060 |
0.1773351252 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Jatropha curcas] |
| 18 |
Hb_024758_010 |
0.1787023318 |
- |
- |
PREDICTED: uncharacterized protein At2g23090-like [Jatropha curcas] |
| 19 |
Hb_002304_040 |
0.179024086 |
- |
- |
PREDICTED: uncharacterized protein LOC105649623 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000085_400 |
0.1794022537 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |