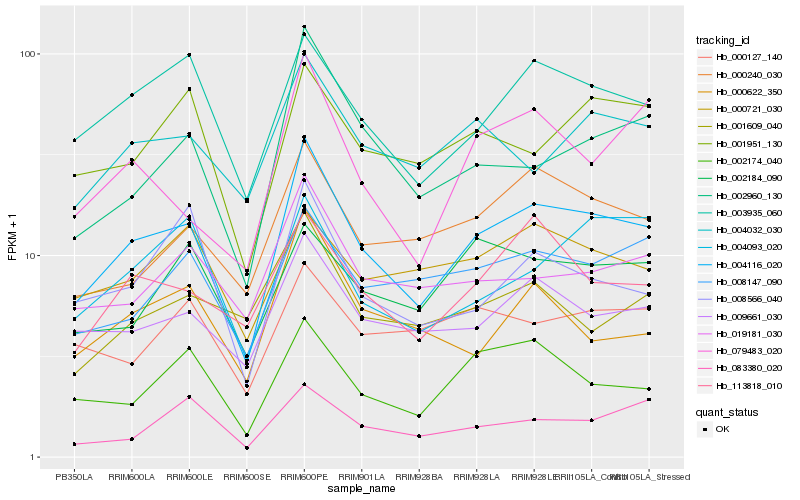
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004116_020 |
0.0 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 2 |
Hb_009661_030 |
0.1222467535 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 3 |
Hb_003935_060 |
0.1238437348 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 4 |
Hb_000240_030 |
0.132532022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002960_130 |
0.1330256947 |
- |
- |
hypothetical protein B456_008G036100 [Gossypium raimondii] |
| 6 |
Hb_113818_010 |
0.1374782764 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 7 |
Hb_019181_030 |
0.1380725319 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 8 |
Hb_002174_040 |
0.1384198556 |
- |
- |
DNAJ heat shock family protein [Theobroma cacao] |
| 9 |
Hb_008147_090 |
0.1402043711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004032_030 |
0.1403684172 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 11 |
Hb_079483_020 |
0.1448274814 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001951_130 |
0.1452942754 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 13 |
Hb_008566_040 |
0.1488193257 |
- |
- |
PREDICTED: uncharacterized protein LOC105647590 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000622_350 |
0.1504679913 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 15 |
Hb_000127_140 |
0.1535645277 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 16 |
Hb_000721_030 |
0.1538938449 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 17 |
Hb_001609_040 |
0.1558262703 |
- |
- |
hypothetical protein POPTR_0017s05650g [Populus trichocarpa] |
| 18 |
Hb_083380_020 |
0.1561038516 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 19 |
Hb_002184_090 |
0.1563193806 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_004093_020 |
0.1563821247 |
- |
- |
Calcium-binding EF-hand family protein [Theobroma cacao] |