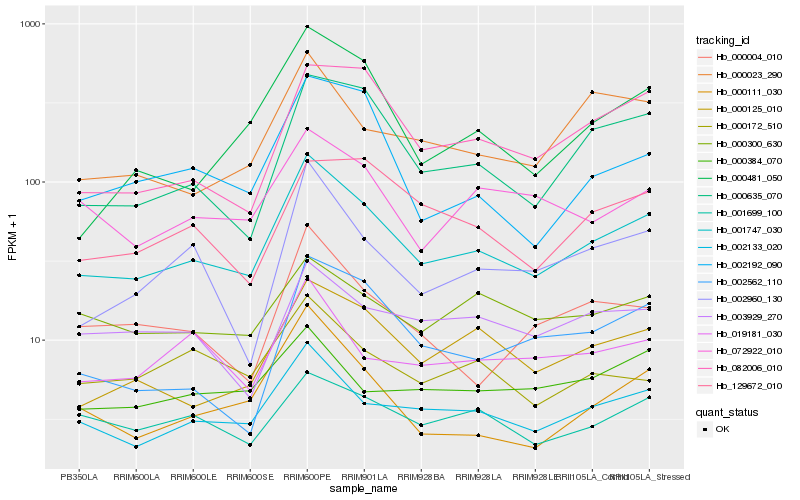
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001747_030 |
0.0 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 2 |
Hb_002133_020 |
0.0879107614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000125_010 |
0.1105122256 |
transcription factor |
TF Family: mTERF |
- |
| 4 |
Hb_000635_070 |
0.1149480821 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 5 |
Hb_072922_010 |
0.1248375696 |
- |
- |
apolipoprotein d, putative [Ricinus communis] |
| 6 |
Hb_000111_030 |
0.1258856556 |
- |
- |
PREDICTED: uncharacterized protein LOC104094030 [Nicotiana tomentosiformis] |
| 7 |
Hb_002960_130 |
0.1268925886 |
- |
- |
hypothetical protein B456_008G036100 [Gossypium raimondii] |
| 8 |
Hb_082006_010 |
0.1271126614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001699_100 |
0.1294603104 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 10 |
Hb_000023_290 |
0.1295371974 |
- |
- |
steroid binding protein, putative [Ricinus communis] |
| 11 |
Hb_019181_030 |
0.1300795199 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 12 |
Hb_002192_090 |
0.1313126603 |
- |
- |
hypothetical protein AMTR_s00029p00055000 [Amborella trichopoda] |
| 13 |
Hb_000004_010 |
0.1344589873 |
- |
- |
hypothetical protein Csa_6G120400 [Cucumis sativus] |
| 14 |
Hb_129672_010 |
0.1351781883 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 15 |
Hb_003929_270 |
0.135349622 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000172_510 |
0.1354994194 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |
| 17 |
Hb_000300_630 |
0.1361408383 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 18 |
Hb_000481_050 |
0.1361603358 |
- |
- |
PREDICTED: uncharacterized protein LOC105636546 [Jatropha curcas] |
| 19 |
Hb_002562_110 |
0.1361644947 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000384_070 |
0.13714967 |
- |
- |
conserved hypothetical protein [Ricinus communis] |