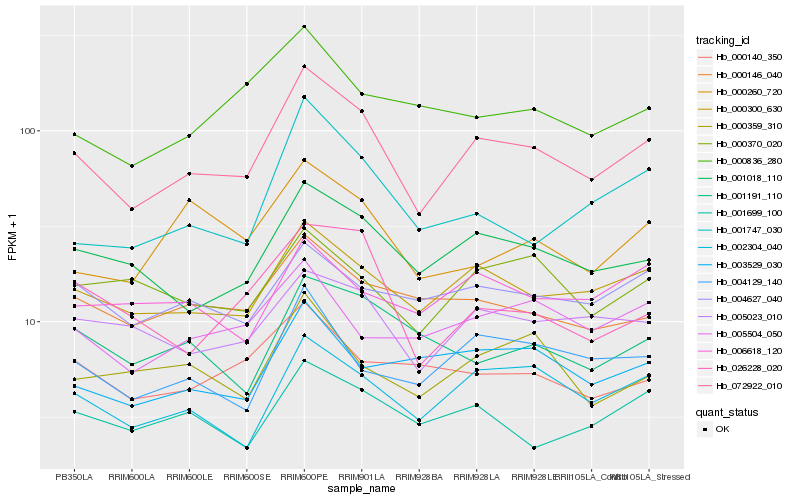
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_072922_010 |
0.0 |
- |
- |
apolipoprotein d, putative [Ricinus communis] |
| 2 |
Hb_000300_630 |
0.0871044317 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 3 |
Hb_001018_110 |
0.0986991346 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_002304_040 |
0.1043540658 |
- |
- |
PREDICTED: uncharacterized protein LOC105649623 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005023_010 |
0.1104573814 |
- |
- |
PREDICTED: putative lipase ROG1 [Jatropha curcas] |
| 6 |
Hb_004129_140 |
0.118106686 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Jatropha curcas] |
| 7 |
Hb_000146_040 |
0.1215931863 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 8 |
Hb_001747_030 |
0.1248375696 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 9 |
Hb_001699_100 |
0.125424899 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 10 |
Hb_026228_020 |
0.1257313874 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 11 |
Hb_000370_020 |
0.1258183986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004627_040 |
0.1266714396 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 13 |
Hb_005504_050 |
0.1281426229 |
- |
- |
putative protein [Arabidopsis thaliana] |
| 14 |
Hb_000359_310 |
0.1292959904 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 15 |
Hb_000260_720 |
0.1296053852 |
- |
- |
PREDICTED: ADP-ribosylation factor-like, partial [Nicotiana tomentosiformis] |
| 16 |
Hb_003529_030 |
0.1304699518 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 17 |
Hb_000140_350 |
0.1310831848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000836_280 |
0.1312155884 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_006618_120 |
0.1324292515 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 20 |
Hb_001191_110 |
0.1336140534 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |