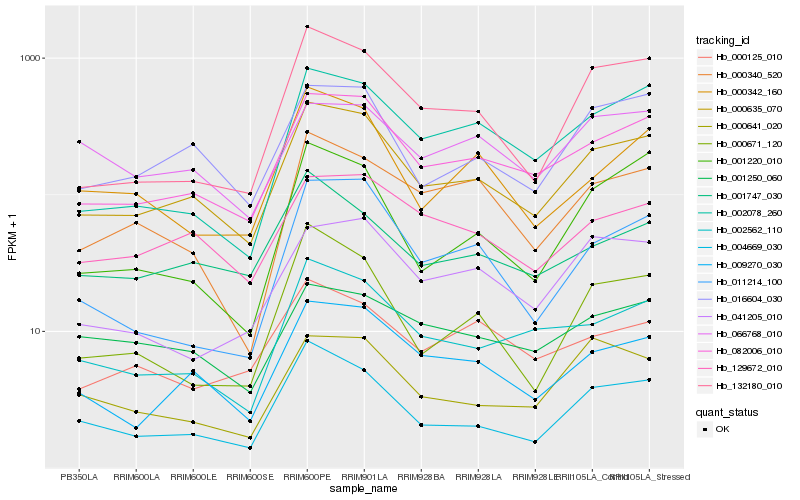
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000635_070 |
0.0 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 2 |
Hb_082006_010 |
0.0574977832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009270_030 |
0.0853359377 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 4 |
Hb_016604_030 |
0.0917682192 |
- |
- |
histone H4 [Zea mays] |
| 5 |
Hb_004669_030 |
0.1010146227 |
- |
- |
- |
| 6 |
Hb_129672_010 |
0.105183049 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 7 |
Hb_002562_110 |
0.1052335589 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001747_030 |
0.1149480821 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 9 |
Hb_132180_010 |
0.115444952 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 10 |
Hb_002078_260 |
0.1190342859 |
- |
- |
unknown [Populus trichocarpa] |
| 11 |
Hb_000340_520 |
0.1225568245 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 12 |
Hb_041205_010 |
0.1241364953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000342_160 |
0.1241927643 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 14 |
Hb_000125_010 |
0.1270206119 |
transcription factor |
TF Family: mTERF |
- |
| 15 |
Hb_001250_060 |
0.1275097453 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 16 |
Hb_000641_020 |
0.1280222268 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 17 |
Hb_011214_100 |
0.1329733755 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 18 |
Hb_000671_120 |
0.1349149547 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 19 |
Hb_001220_010 |
0.1358528945 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 20 |
Hb_066768_010 |
0.1362258852 |
- |
- |
PREDICTED: outer envelope pore protein 16-3, chloroplastic/mitochondrial [Jatropha curcas] |