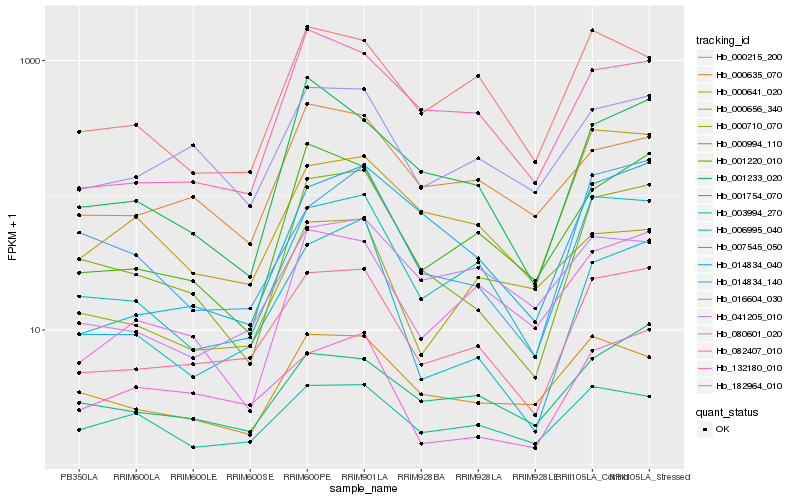
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_182964_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |
| 2 |
Hb_006995_040 |
0.1002780634 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 3 |
Hb_016604_030 |
0.1223116815 |
- |
- |
histone H4 [Zea mays] |
| 4 |
Hb_001220_010 |
0.137987774 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 5 |
Hb_007545_050 |
0.1384686599 |
- |
- |
- |
| 6 |
Hb_000994_110 |
0.1390331708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001754_070 |
0.14103574 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 8 |
Hb_000656_340 |
0.1472352988 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 9 |
Hb_080601_020 |
0.1492784918 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 10 |
Hb_132180_010 |
0.1506218879 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 11 |
Hb_000215_200 |
0.1515781074 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 12 |
Hb_082407_010 |
0.1536821761 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 13 |
Hb_000710_070 |
0.1540084597 |
- |
- |
PREDICTED: uncharacterized protein LOC105645553 [Jatropha curcas] |
| 14 |
Hb_003994_270 |
0.1548269442 |
- |
- |
PREDICTED: uncharacterized protein LOC105646152 [Jatropha curcas] |
| 15 |
Hb_000635_070 |
0.1562050152 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 16 |
Hb_014834_140 |
0.159393397 |
- |
- |
Uncharacterized protein TCM_015942 [Theobroma cacao] |
| 17 |
Hb_001233_020 |
0.1595078637 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 18 |
Hb_041205_010 |
0.1596391962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_014834_040 |
0.1601089564 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 20 |
Hb_000641_020 |
0.1605391331 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |