| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
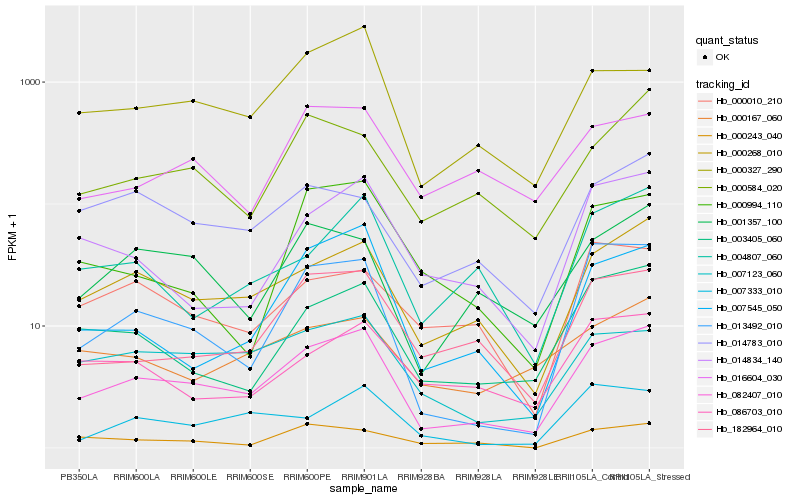
Hb_082407_010 |
0.0 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 2 |
Hb_000268_010 |
0.1149341093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000327_290 |
0.1447083503 |
- |
- |
Histone H4 [Triticum urartu] |
| 4 |
Hb_013492_010 |
0.1458023549 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 5 |
Hb_003405_060 |
0.149236294 |
- |
- |
- |
| 6 |
Hb_182964_010 |
0.1536821761 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |
| 7 |
Hb_007123_060 |
0.1540134042 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 8 |
Hb_007545_050 |
0.1575404739 |
- |
- |
- |
| 9 |
Hb_001357_100 |
0.1605240025 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 10 |
Hb_014834_140 |
0.1659337312 |
- |
- |
Uncharacterized protein TCM_015942 [Theobroma cacao] |
| 11 |
Hb_000584_020 |
0.1694516233 |
- |
- |
hypothetical protein PHAVU_002G233500g, partial [Phaseolus vulgaris] |
| 12 |
Hb_004807_060 |
0.1703497423 |
- |
- |
- |
| 13 |
Hb_014783_010 |
0.1704211467 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_007333_010 |
0.1718472911 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000243_040 |
0.1750118941 |
- |
- |
- |
| 16 |
Hb_086703_010 |
0.1762878157 |
- |
- |
PREDICTED: F-box protein At5g52880 [Jatropha curcas] |
| 17 |
Hb_000167_060 |
0.1785630641 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 31 kDa protein [Jatropha curcas] |
| 18 |
Hb_016604_030 |
0.1793785539 |
- |
- |
histone H4 [Zea mays] |
| 19 |
Hb_000010_210 |
0.1815906183 |
- |
- |
PREDICTED: centromere protein X [Jatropha curcas] |
| 20 |
Hb_000994_110 |
0.1827803904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |