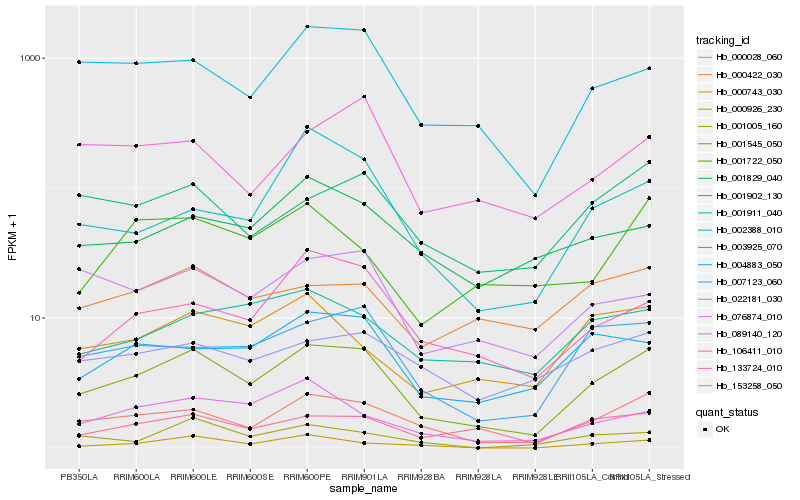
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001545_050 |
0.0 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 2 |
Hb_000028_060 |
0.1076920763 |
- |
- |
- |
| 3 |
Hb_007123_060 |
0.1408703992 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 4 |
Hb_003925_070 |
0.147026236 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 5 |
Hb_153258_050 |
0.1533633723 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 6 |
Hb_001902_130 |
0.1605709272 |
- |
- |
60S ribosomal protein L36-2 [Morus notabilis] |
| 7 |
Hb_004883_050 |
0.1609128002 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 8 |
Hb_133724_010 |
0.1661667356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_076874_010 |
0.1686823518 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 10 |
Hb_089140_120 |
0.1701030047 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 11 |
Hb_000743_030 |
0.1733604084 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 12 |
Hb_001911_040 |
0.1752131439 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 13 |
Hb_022181_030 |
0.177202431 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 14 |
Hb_106411_010 |
0.1778437247 |
- |
- |
Ribosomal protein S25 family protein [Theobroma cacao] |
| 15 |
Hb_000422_030 |
0.1828693305 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 16 |
Hb_001829_040 |
0.1835844429 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 17 |
Hb_001005_160 |
0.1849012536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001722_050 |
0.186759508 |
- |
- |
calcium binding family protein [Populus trichocarpa] |
| 19 |
Hb_000926_230 |
0.1877649957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002388_010 |
0.1878740936 |
- |
- |
- |