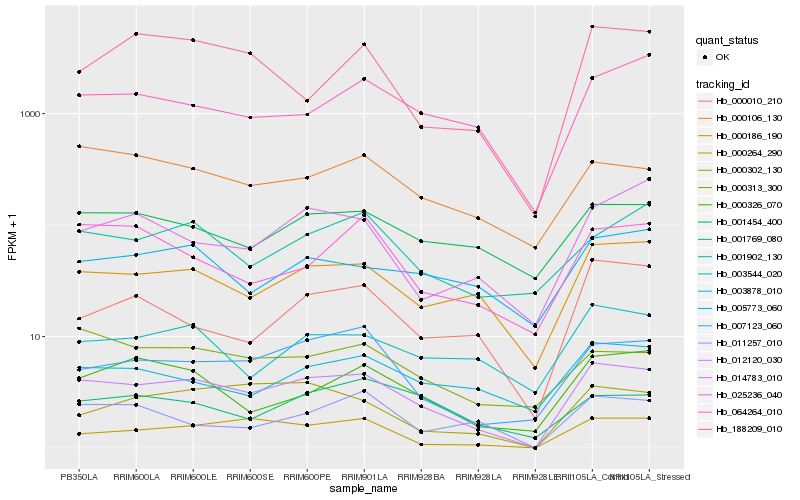
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012120_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000186_190 |
0.1352751216 |
- |
- |
PREDICTED: uncharacterized protein At4g22160 [Jatropha curcas] |
| 3 |
Hb_007123_060 |
0.1459816291 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 4 |
Hb_000264_290 |
0.1544617499 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 5 |
Hb_000106_130 |
0.1642032209 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000302_130 |
0.164345701 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 7 |
Hb_064264_010 |
0.1645275402 |
- |
- |
hypothetical protein [Zea mays] |
| 8 |
Hb_000326_070 |
0.1654763083 |
- |
- |
phenylalanyl-tRNA synthetase beta chain, putative [Ricinus communis] |
| 9 |
Hb_000313_300 |
0.1659358222 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Jatropha curcas] |
| 10 |
Hb_001454_400 |
0.1662758556 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic [Jatropha curcas] |
| 11 |
Hb_003544_020 |
0.1701487165 |
- |
- |
PREDICTED: elongin-A [Jatropha curcas] |
| 12 |
Hb_003878_010 |
0.1713166814 |
- |
- |
PREDICTED: uncharacterized protein LOC105632145 [Jatropha curcas] |
| 13 |
Hb_000010_210 |
0.1722738714 |
- |
- |
PREDICTED: centromere protein X [Jatropha curcas] |
| 14 |
Hb_001902_130 |
0.1723798213 |
- |
- |
60S ribosomal protein L36-2 [Morus notabilis] |
| 15 |
Hb_005773_060 |
0.172430656 |
- |
- |
defender against cell death, putative [Ricinus communis] |
| 16 |
Hb_011257_010 |
0.1728148353 |
- |
- |
- |
| 17 |
Hb_188209_010 |
0.1753576336 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_025236_040 |
0.1761932125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001769_080 |
0.1768457992 |
- |
- |
hypothetical protein SORBIDRAFT_10g023200 [Sorghum bicolor] |
| 20 |
Hb_014783_010 |
0.1783039011 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |