| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
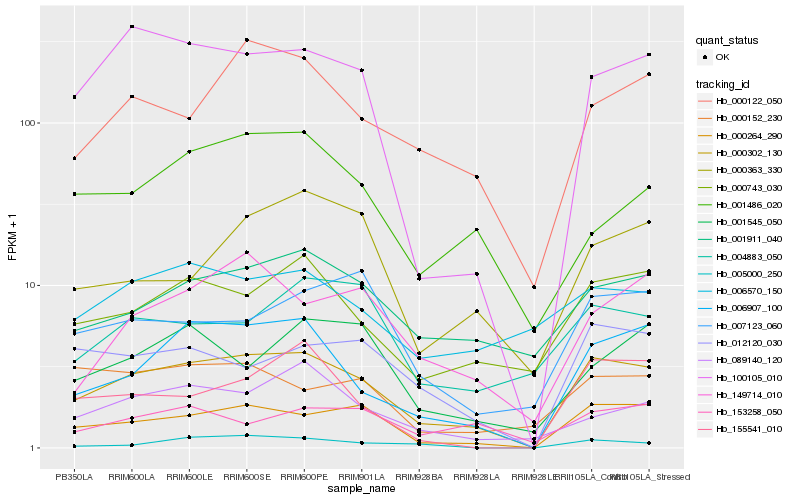
Hb_000302_130 |
0.0 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 2 |
Hb_000264_290 |
0.1202001146 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 3 |
Hb_006907_100 |
0.1375355254 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 4 |
Hb_100105_010 |
0.1548585384 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 5 |
Hb_000743_030 |
0.1554355187 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 6 |
Hb_000122_050 |
0.1588628161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_012120_030 |
0.164345701 |
- |
- |
- |
| 8 |
Hb_001911_040 |
0.1688384721 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 9 |
Hb_149714_010 |
0.1820864558 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 10 |
Hb_089140_120 |
0.1825747985 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 11 |
Hb_007123_060 |
0.1857136625 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 12 |
Hb_001545_050 |
0.1891751656 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 13 |
Hb_155541_010 |
0.1904302063 |
- |
- |
- |
| 14 |
Hb_006570_150 |
0.1909637679 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000363_330 |
0.1919495022 |
- |
- |
- |
| 16 |
Hb_001486_020 |
0.1928727125 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 17 |
Hb_000152_230 |
0.1931241598 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 18 |
Hb_004883_050 |
0.1937711331 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 19 |
Hb_153258_050 |
0.1939317732 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 20 |
Hb_005000_250 |
0.1960192137 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 4, mitochondrial [Vitis vinifera] |