| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
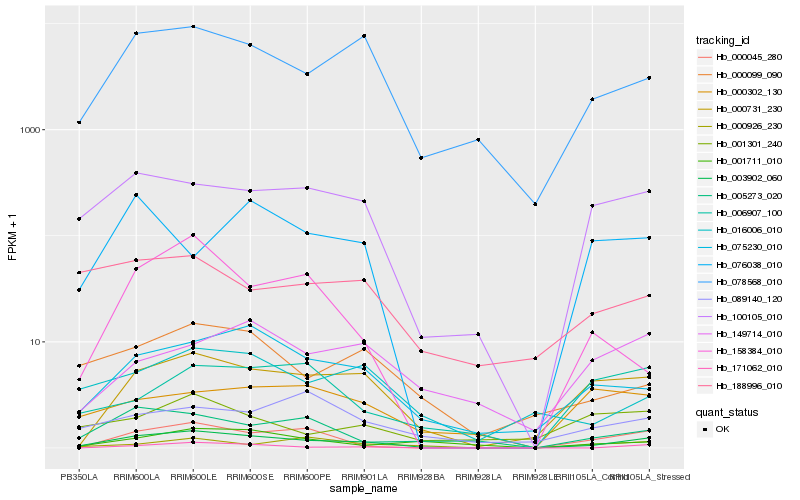
Hb_003902_060 |
0.0 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 2 |
Hb_078568_010 |
0.1944594469 |
- |
- |
hypothetical protein PRUPE_ppa013245mg [Prunus persica] |
| 3 |
Hb_100105_010 |
0.1945126104 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 4 |
Hb_000731_230 |
0.1955745294 |
- |
- |
PREDICTED: abnormal spindle-like microcephaly-associated protein homolog [Jatropha curcas] |
| 5 |
Hb_075230_010 |
0.2072556693 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like [Glycine max] |
| 6 |
Hb_000099_090 |
0.2331210819 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 7 |
Hb_000045_280 |
0.2347911531 |
- |
- |
hypothetical protein JCGZ_04076 [Jatropha curcas] |
| 8 |
Hb_171062_010 |
0.2365297987 |
- |
- |
hypothetical protein VITISV_036059 [Vitis vinifera] |
| 9 |
Hb_005273_020 |
0.2373744475 |
- |
- |
- |
| 10 |
Hb_016006_010 |
0.2387092334 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_001711_010 |
0.2544970441 |
- |
- |
- |
| 12 |
Hb_006907_100 |
0.2570992987 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 13 |
Hb_149714_010 |
0.2602419539 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 14 |
Hb_188996_010 |
0.2626204204 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000926_230 |
0.2627671651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_089140_120 |
0.2646978851 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 17 |
Hb_000302_130 |
0.2661332611 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_001301_240 |
0.2702175428 |
- |
- |
- |
| 19 |
Hb_076038_010 |
0.2712854689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_158384_010 |
0.2718869958 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |