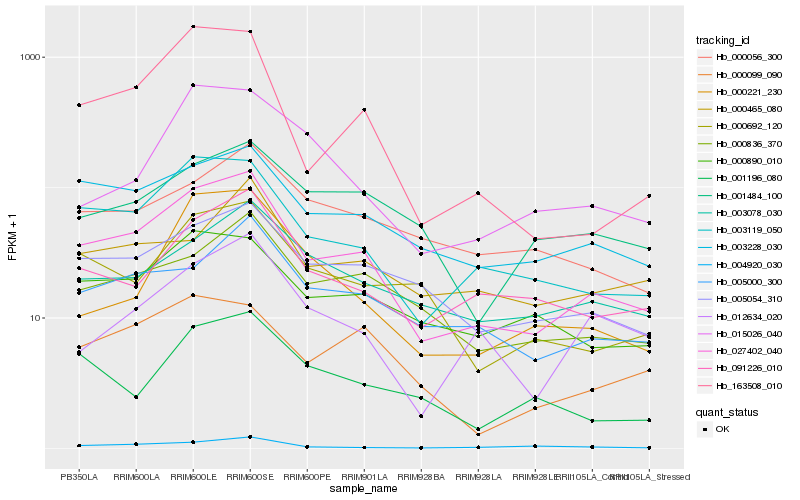
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027402_040 |
0.0 |
- |
- |
Thioredoxin II, putative [Ricinus communis] |
| 2 |
Hb_003119_050 |
0.1077088721 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_003228_030 |
0.1365200043 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_005054_310 |
0.1411277353 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 5 |
Hb_163508_010 |
0.1451966292 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 6 |
Hb_000836_370 |
0.1476393273 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_005000_300 |
0.1503646866 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 8 |
Hb_091226_010 |
0.1509932038 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 9 |
Hb_000890_010 |
0.155399845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000692_120 |
0.1557749967 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 11 |
Hb_003078_030 |
0.1649152442 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 12 |
Hb_000099_090 |
0.1664795656 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 13 |
Hb_004920_030 |
0.171653083 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase CKI1 [Jatropha curcas] |
| 14 |
Hb_001196_080 |
0.1733229903 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 15 |
Hb_012634_020 |
0.1734241779 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000056_300 |
0.1738486786 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000221_230 |
0.1757046123 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 18 |
Hb_001484_100 |
0.1771119645 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 19 |
Hb_015026_040 |
0.1773310627 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 20 |
Hb_000465_080 |
0.1780693374 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |