| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
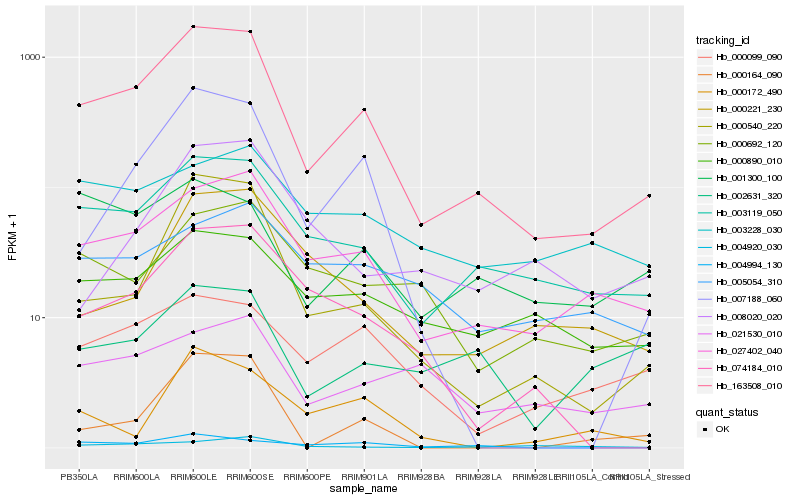
Hb_163508_010 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 2 |
Hb_027402_040 |
0.1451966292 |
- |
- |
Thioredoxin II, putative [Ricinus communis] |
| 3 |
Hb_003119_050 |
0.1564366304 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000540_220 |
0.1751270526 |
- |
- |
hypothetical protein JCGZ_05605 [Jatropha curcas] |
| 5 |
Hb_007188_060 |
0.1989303297 |
- |
- |
- |
| 6 |
Hb_074184_010 |
0.204694763 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 7 |
Hb_000099_090 |
0.2152045795 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 8 |
Hb_000890_010 |
0.2191276842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000692_120 |
0.2203542009 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 10 |
Hb_000221_230 |
0.2242618503 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 11 |
Hb_000164_090 |
0.2268389037 |
- |
- |
hypothetical protein [Plasmodium berghei strain ANKA], partial [Plasmodium berghei ANKA] |
| 12 |
Hb_004994_130 |
0.2286795741 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |
| 13 |
Hb_001300_100 |
0.234266084 |
- |
- |
- |
| 14 |
Hb_021530_010 |
0.2402504155 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 15 |
Hb_003228_030 |
0.2404107553 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_000172_490 |
0.2417996489 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 17 |
Hb_004920_030 |
0.2428511361 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase CKI1 [Jatropha curcas] |
| 18 |
Hb_005054_310 |
0.2436146123 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 19 |
Hb_008020_020 |
0.2438458763 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002631_320 |
0.2444380234 |
- |
- |
hypothetical protein JCGZ_22628 [Jatropha curcas] |