| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
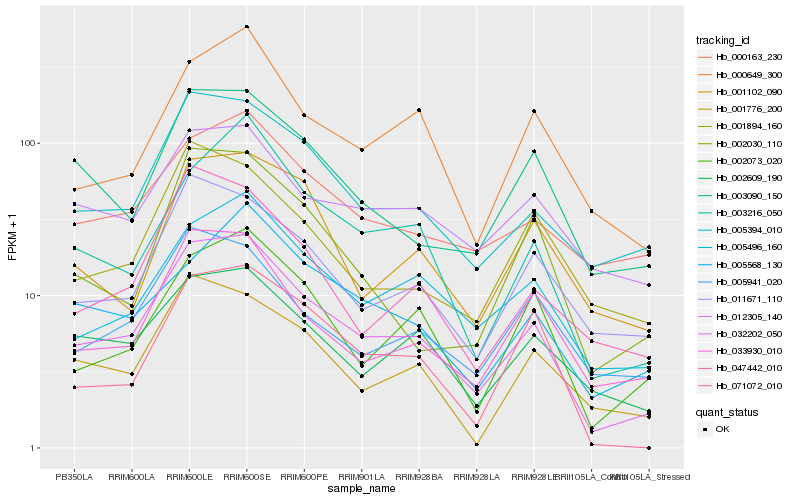
Hb_032202_050 |
0.0 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005394_010 |
0.1149216148 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 3 |
Hb_005496_160 |
0.127407826 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_033930_010 |
0.1286726553 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 5 |
Hb_011671_110 |
0.1331934988 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 6 |
Hb_002609_190 |
0.1362771976 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 7 |
Hb_003216_050 |
0.1372894993 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 8 |
Hb_000649_300 |
0.1383728033 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 9 |
Hb_002073_020 |
0.1387914567 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 10 |
Hb_047442_010 |
0.1390508915 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 11 |
Hb_003090_150 |
0.1393629517 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 12 |
Hb_071072_010 |
0.1434643587 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_001776_200 |
0.1436170685 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 14 |
Hb_012305_140 |
0.1439111014 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000163_230 |
0.1441934982 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 16 |
Hb_005941_020 |
0.1461756978 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 17 |
Hb_002030_110 |
0.149635153 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 18 |
Hb_001894_160 |
0.1516999796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001102_090 |
0.1518509317 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 20 |
Hb_005568_130 |
0.1546490422 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |