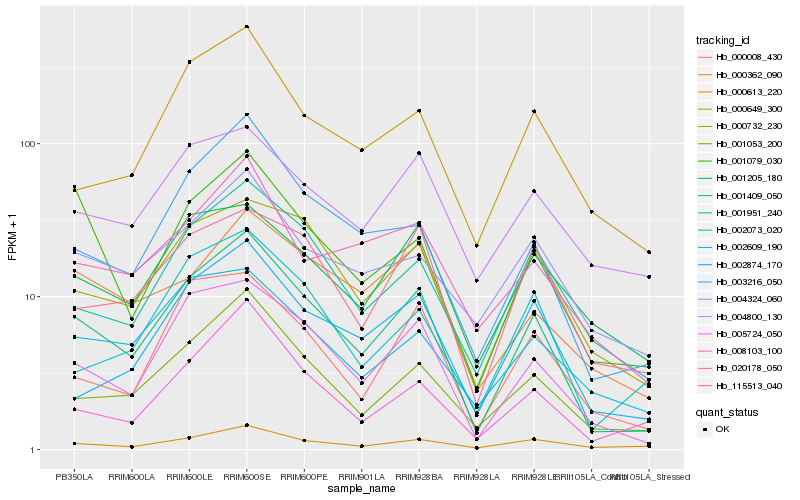
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001409_050 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_005724_050 |
0.1004003851 |
- |
- |
- |
| 3 |
Hb_001053_200 |
0.1233715557 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 4 |
Hb_003216_050 |
0.1275098766 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 5 |
Hb_000732_230 |
0.1378789675 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001951_240 |
0.1386929024 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 7 |
Hb_000008_430 |
0.1464381433 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008103_100 |
0.1509390083 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 9 |
Hb_000649_300 |
0.151593514 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 10 |
Hb_001079_030 |
0.1527201746 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Jatropha curcas] |
| 11 |
Hb_020178_050 |
0.1551213544 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 12 |
Hb_000613_220 |
0.1553973355 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 13 |
Hb_000362_090 |
0.1554482996 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 14 |
Hb_115513_040 |
0.1561901431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002874_170 |
0.1563037326 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_004324_060 |
0.157850943 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 17 |
Hb_002609_190 |
0.1579845932 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 18 |
Hb_002073_020 |
0.1587167406 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 19 |
Hb_001205_180 |
0.1599095398 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 20 |
Hb_004800_130 |
0.1648187217 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |