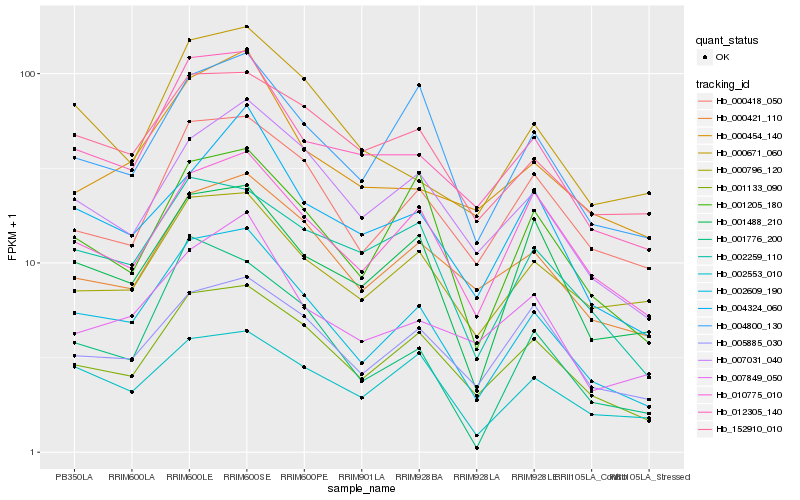
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002609_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 2 |
Hb_001133_090 |
0.0933871743 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 3 |
Hb_012305_140 |
0.0992848688 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_004800_130 |
0.1137499689 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 5 |
Hb_007031_040 |
0.1150584822 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 6 |
Hb_002259_110 |
0.1171359929 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 7 |
Hb_001205_180 |
0.1179642141 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 8 |
Hb_001488_210 |
0.1191639434 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |
| 9 |
Hb_000418_050 |
0.1200569474 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 10 |
Hb_000421_110 |
0.12013506 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002553_010 |
0.121560618 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 2-like [Populus euphratica] |
| 12 |
Hb_010775_010 |
0.1241179159 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007849_050 |
0.1244157529 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 14 |
Hb_004324_060 |
0.1248780762 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 15 |
Hb_152910_010 |
0.1256746405 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 16 |
Hb_001776_200 |
0.1262523391 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 17 |
Hb_000454_140 |
0.1274044875 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 18 |
Hb_000671_060 |
0.129580583 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_005885_030 |
0.129934201 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 20 |
Hb_000796_120 |
0.1304567275 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |