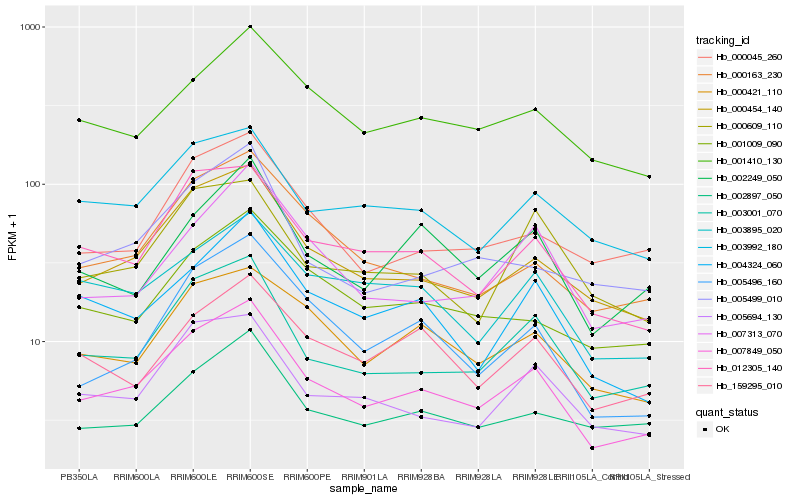
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007849_050 |
0.0 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 2 |
Hb_000454_140 |
0.0750113397 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 3 |
Hb_003001_070 |
0.0776588446 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0002s24750g [Populus trichocarpa] |
| 4 |
Hb_012305_140 |
0.0916878017 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_005694_130 |
0.0993580183 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 6 |
Hb_000163_230 |
0.1032706542 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 7 |
Hb_004324_060 |
0.1055941165 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 8 |
Hb_000421_110 |
0.1069882292 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003992_180 |
0.1071105642 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005496_160 |
0.108579735 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001410_130 |
0.108623573 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 12 |
Hb_000045_260 |
0.1111756667 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_007313_070 |
0.1123131007 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 14 |
Hb_003895_020 |
0.1131307573 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001009_090 |
0.1144555922 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 16 |
Hb_002249_050 |
0.1154290792 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 17 |
Hb_002897_050 |
0.116220974 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 18 |
Hb_000609_110 |
0.1183963861 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 19 |
Hb_159295_010 |
0.1186153819 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 20 |
Hb_005499_010 |
0.1190932167 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |