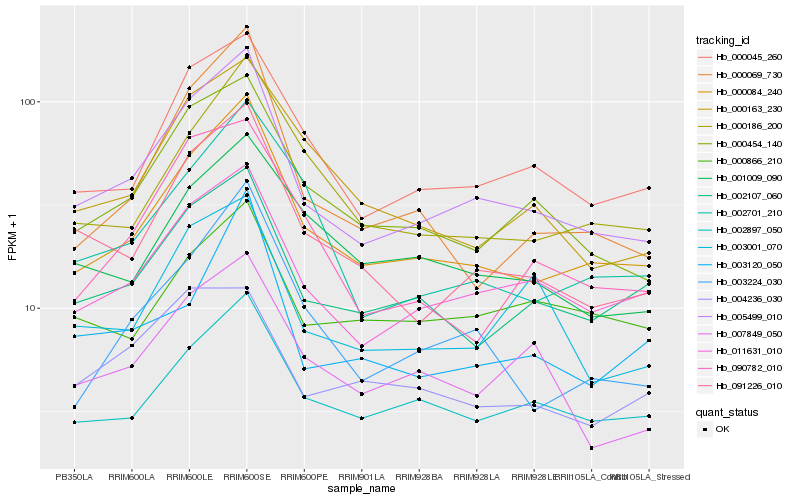
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005499_010 |
0.0 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 2 |
Hb_000084_240 |
0.0710173887 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 3 |
Hb_011631_010 |
0.0814803477 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 4 |
Hb_091226_010 |
0.0898794173 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 5 |
Hb_000454_140 |
0.0910433017 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 6 |
Hb_000045_260 |
0.0933125737 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_002897_050 |
0.1060877423 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 8 |
Hb_002107_060 |
0.107407674 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 9 |
Hb_003001_070 |
0.1156472353 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0002s24750g [Populus trichocarpa] |
| 10 |
Hb_007849_050 |
0.1190932167 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 11 |
Hb_000069_730 |
0.1224105083 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000163_230 |
0.1240821579 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 13 |
Hb_000186_200 |
0.125342744 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 14 |
Hb_003224_030 |
0.1254915291 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 15 |
Hb_090782_010 |
0.1314839437 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 16 |
Hb_002701_210 |
0.1317122673 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 17 |
Hb_001009_090 |
0.1320016159 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 18 |
Hb_003120_050 |
0.1335693778 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 19 |
Hb_000866_210 |
0.1367022451 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004236_030 |
0.1367103292 |
- |
- |
unknown [Lotus japonicus] |