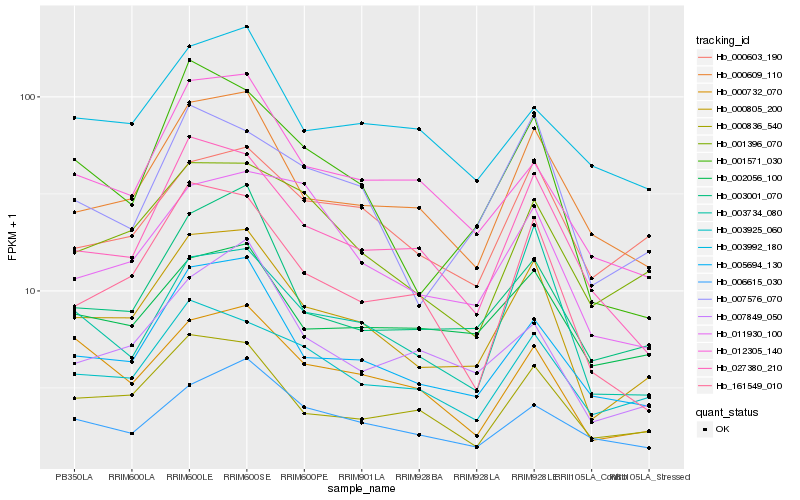
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000805_200 |
0.0 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005694_130 |
0.0896518336 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 3 |
Hb_000609_110 |
0.1004907644 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 4 |
Hb_000836_540 |
0.1054011598 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 5 |
Hb_027380_210 |
0.1091052027 |
- |
- |
PREDICTED: uncharacterized protein LOC105634030 [Jatropha curcas] |
| 6 |
Hb_003001_070 |
0.1113894449 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0002s24750g [Populus trichocarpa] |
| 7 |
Hb_001571_030 |
0.112966837 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007576_070 |
0.1150068439 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X3 [Jatropha curcas] |
| 9 |
Hb_161549_010 |
0.1150743278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001396_070 |
0.1163933706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003925_060 |
0.1166836789 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000732_070 |
0.1209439746 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 13 |
Hb_012305_140 |
0.1213245817 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002056_100 |
0.1226461548 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 15 |
Hb_007849_050 |
0.1243571775 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 16 |
Hb_000603_190 |
0.1289045725 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 17 |
Hb_003734_080 |
0.1307347999 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 18 |
Hb_011930_100 |
0.1333810049 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 19 |
Hb_006615_030 |
0.1348813157 |
- |
- |
prolyl oligopeptidase family protein [Populus trichocarpa] |
| 20 |
Hb_003992_180 |
0.1352456707 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |