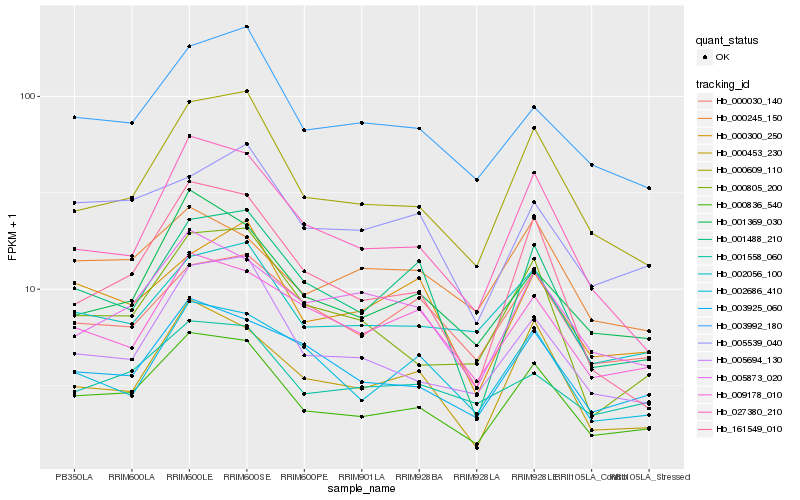
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_540 |
0.0 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 2 |
Hb_000609_110 |
0.0774370859 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 3 |
Hb_005694_130 |
0.0938814069 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 4 |
Hb_001369_030 |
0.094716131 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 5 |
Hb_003925_060 |
0.0955115396 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000453_230 |
0.0980533531 |
- |
- |
PREDICTED: NO-associated protein 1, chloroplastic/mitochondrial [Jatropha curcas] |
| 7 |
Hb_002056_100 |
0.0988186334 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 8 |
Hb_003992_180 |
0.0988932493 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 9 |
Hb_027380_210 |
0.0999809903 |
- |
- |
PREDICTED: uncharacterized protein LOC105634030 [Jatropha curcas] |
| 10 |
Hb_001558_060 |
0.1052899324 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000805_200 |
0.1054011598 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_009178_010 |
0.109916218 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 13 |
Hb_005873_020 |
0.110511861 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 14 |
Hb_002686_410 |
0.1138440304 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000030_140 |
0.1142441822 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001488_210 |
0.1146771159 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |
| 17 |
Hb_000300_250 |
0.1164561725 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 18 |
Hb_161549_010 |
0.1184492548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005539_040 |
0.1184969573 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000245_150 |
0.1193037276 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |