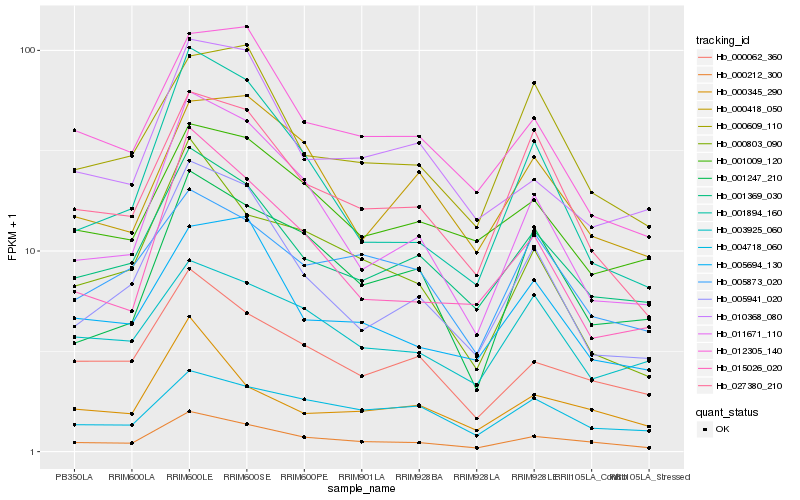
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_300 |
0.0 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 2 |
Hb_001369_030 |
0.0836259936 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 3 |
Hb_000062_360 |
0.0885733754 |
- |
- |
PREDICTED: DNA polymerase alpha subunit B [Jatropha curcas] |
| 4 |
Hb_027380_210 |
0.1015010972 |
- |
- |
PREDICTED: uncharacterized protein LOC105634030 [Jatropha curcas] |
| 5 |
Hb_011671_110 |
0.1075399255 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 6 |
Hb_015026_020 |
0.1147695636 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 7 |
Hb_005941_020 |
0.1157097744 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 8 |
Hb_012305_140 |
0.1195095808 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_005694_130 |
0.1197457476 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 10 |
Hb_001894_160 |
0.1211053911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001009_120 |
0.1221633332 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000345_290 |
0.1226565132 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_000803_090 |
0.1228490843 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |
| 14 |
Hb_001247_210 |
0.1241366149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004718_060 |
0.1247047605 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000609_110 |
0.1264770836 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 17 |
Hb_005873_020 |
0.1280748717 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 18 |
Hb_010368_080 |
0.1283655895 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 19 |
Hb_003925_060 |
0.1295613892 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000418_050 |
0.1304211768 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |