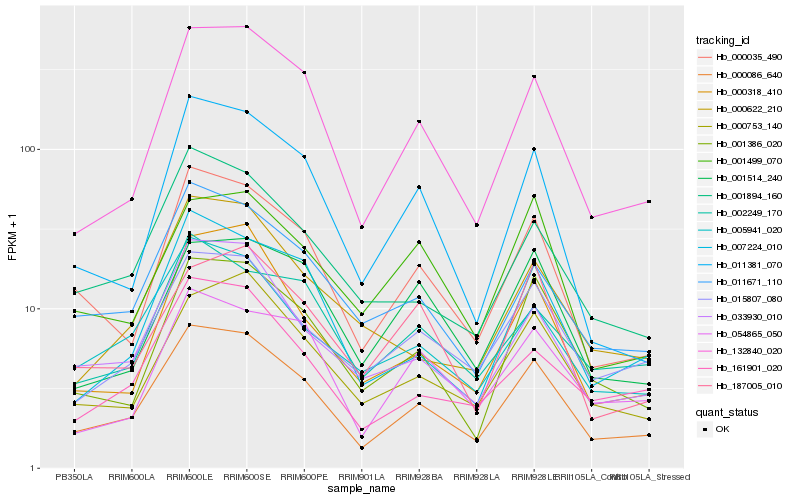
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_640 |
0.0 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 2 |
Hb_000035_490 |
0.0887816394 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 3 |
Hb_132840_020 |
0.0908853842 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 4 |
Hb_015807_080 |
0.0991471387 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 5 |
Hb_033930_010 |
0.1024243373 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 6 |
Hb_005941_020 |
0.1058399133 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 7 |
Hb_000753_140 |
0.1070502282 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 8 |
Hb_001386_020 |
0.1072680256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011671_110 |
0.109059241 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 10 |
Hb_001894_160 |
0.1097650532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007224_010 |
0.1126321497 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 12 |
Hb_161901_020 |
0.1145182919 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 13 |
Hb_000318_410 |
0.1160385125 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 14 |
Hb_011381_070 |
0.124908972 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 15 |
Hb_054865_050 |
0.1255662833 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_002249_170 |
0.129643366 |
- |
- |
copine, putative [Ricinus communis] |
| 17 |
Hb_000622_210 |
0.1311521671 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 18 |
Hb_001514_240 |
0.1358921092 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 19 |
Hb_187005_010 |
0.1367724836 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 20 |
Hb_001499_070 |
0.1371871766 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |