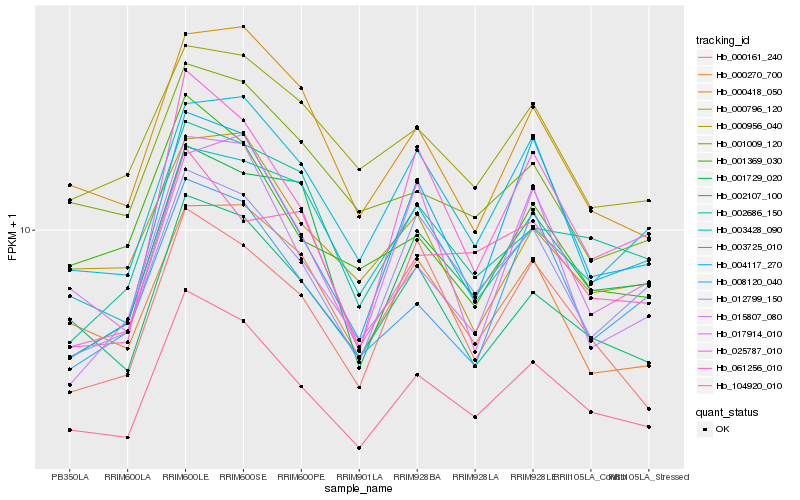
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_104920_010 |
0.0 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 2 |
Hb_002107_100 |
0.0878460577 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 3 |
Hb_012799_150 |
0.0994100731 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methyl-CpG-binding domain-containing protein 13 [Jatropha curcas] |
| 4 |
Hb_025787_010 |
0.1002976909 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 5 |
Hb_004117_270 |
0.1080573184 |
- |
- |
- |
| 6 |
Hb_000418_050 |
0.1084794866 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 7 |
Hb_001369_030 |
0.1098955128 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 8 |
Hb_003725_010 |
0.1102092151 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 9 |
Hb_001729_020 |
0.1106075481 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000270_700 |
0.1118078915 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 11 |
Hb_000161_240 |
0.1121682039 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_015807_080 |
0.1146305224 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 13 |
Hb_002686_150 |
0.1151143156 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_008120_040 |
0.1183830632 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 15 |
Hb_000796_120 |
0.1216086519 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 16 |
Hb_003428_090 |
0.1224614092 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 17 |
Hb_017914_010 |
0.1231676586 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 18 |
Hb_000956_040 |
0.1236066881 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 19 |
Hb_061256_010 |
0.1237942092 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001009_120 |
0.1239436878 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |