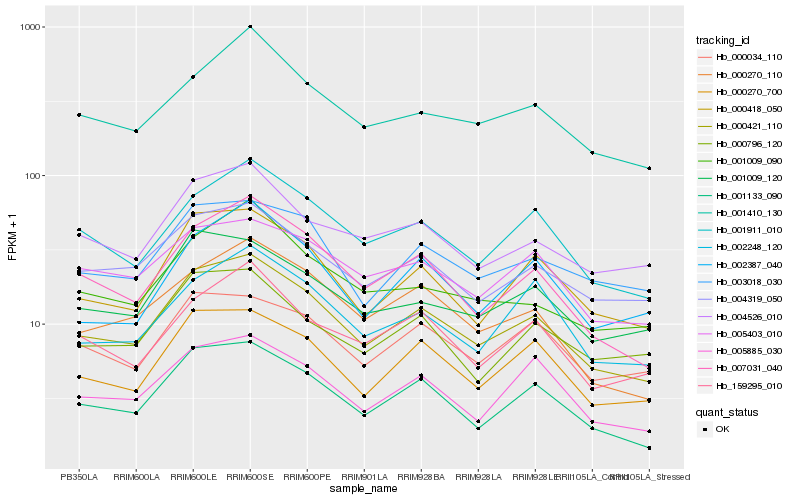
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000421_110 |
0.0 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001133_090 |
0.0575553921 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 3 |
Hb_007031_040 |
0.0666873318 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 4 |
Hb_000418_050 |
0.0678585073 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 5 |
Hb_000270_700 |
0.0717490459 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 6 |
Hb_001911_010 |
0.0747550519 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 7 |
Hb_004319_050 |
0.077683699 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 8 |
Hb_000270_110 |
0.0825721602 |
- |
- |
PREDICTED: calcium-dependent protein kinase 10 [Jatropha curcas] |
| 9 |
Hb_003018_030 |
0.0843917162 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 10 |
Hb_004526_010 |
0.0854549953 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_005885_030 |
0.086001715 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 12 |
Hb_001009_120 |
0.0870119694 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001410_130 |
0.0876290829 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 14 |
Hb_159295_010 |
0.0883854315 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 15 |
Hb_005403_010 |
0.0904493005 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 16 |
Hb_002387_040 |
0.0905973038 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 17 |
Hb_000034_110 |
0.090711681 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 18 |
Hb_002248_120 |
0.0938647247 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 19 |
Hb_001009_090 |
0.0942132748 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 20 |
Hb_000796_120 |
0.0949418598 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |