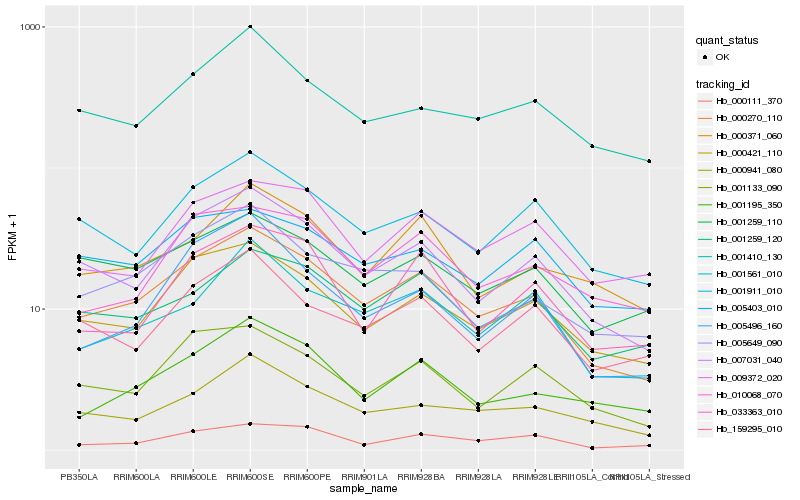
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_110 |
0.0 |
- |
- |
PREDICTED: calcium-dependent protein kinase 10 [Jatropha curcas] |
| 2 |
Hb_007031_040 |
0.0705812516 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 3 |
Hb_000421_110 |
0.0825721602 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001133_090 |
0.1008978901 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 5 |
Hb_005649_090 |
0.1022720116 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 6 |
Hb_001911_010 |
0.1025970574 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 7 |
Hb_000941_080 |
0.1042497117 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X3 [Populus euphratica] |
| 8 |
Hb_001561_010 |
0.1050114379 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001259_120 |
0.106835867 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 10 |
Hb_001259_110 |
0.1111978515 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 3 [Jatropha curcas] |
| 11 |
Hb_001410_130 |
0.1117478293 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 12 |
Hb_000371_060 |
0.1133712959 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 13 |
Hb_159295_010 |
0.1138011801 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 14 |
Hb_001195_350 |
0.1143530781 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 15 |
Hb_000111_370 |
0.1151038857 |
- |
- |
PREDICTED: cation/H(+) antiporter 2-like [Jatropha curcas] |
| 16 |
Hb_010068_070 |
0.1152673783 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 17 |
Hb_005403_010 |
0.1175361174 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 18 |
Hb_009372_020 |
0.1189592765 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 19 |
Hb_005496_160 |
0.1197841991 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_033363_010 |
0.1199884046 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |