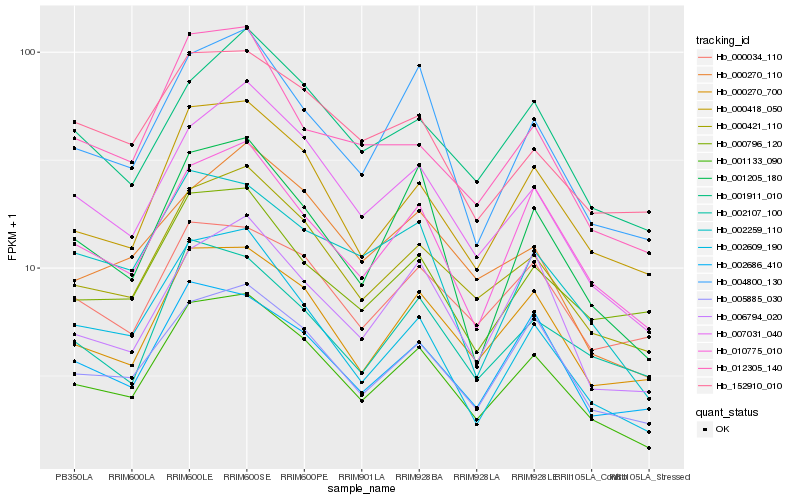
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001133_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 2 |
Hb_000421_110 |
0.0575553921 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000418_050 |
0.064890977 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 4 |
Hb_007031_040 |
0.0705692885 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 5 |
Hb_005885_030 |
0.0713637816 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 6 |
Hb_000270_700 |
0.0724089287 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 7 |
Hb_010775_010 |
0.0762837303 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004800_130 |
0.0807864237 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 9 |
Hb_001205_180 |
0.0897903649 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 10 |
Hb_002686_410 |
0.0914467789 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_002609_190 |
0.0933871743 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 12 |
Hb_001911_010 |
0.0939077638 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 13 |
Hb_006794_020 |
0.0943511385 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 14 |
Hb_002259_110 |
0.0979610358 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 15 |
Hb_002107_100 |
0.099692325 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_000270_110 |
0.1008978901 |
- |
- |
PREDICTED: calcium-dependent protein kinase 10 [Jatropha curcas] |
| 17 |
Hb_000796_120 |
0.1015525378 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_152910_010 |
0.1020025088 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 19 |
Hb_000034_110 |
0.1022955359 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 20 |
Hb_012305_140 |
0.1029364591 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |