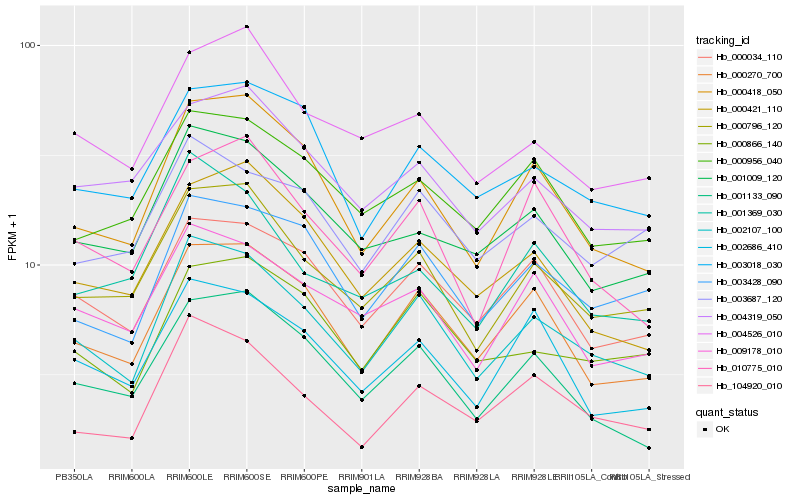
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002107_100 |
0.0 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 2 |
Hb_000796_120 |
0.075565972 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000270_700 |
0.0797805985 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 4 |
Hb_000418_050 |
0.0805029809 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 5 |
Hb_104920_010 |
0.0878460577 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 6 |
Hb_000034_110 |
0.088776266 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 7 |
Hb_003428_090 |
0.0887778454 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 8 |
Hb_000866_140 |
0.0937458602 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 9 |
Hb_009178_010 |
0.0969372629 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 10 |
Hb_001009_120 |
0.0983950572 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001133_090 |
0.099692325 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 12 |
Hb_003018_030 |
0.1004227801 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 13 |
Hb_002686_410 |
0.1008434398 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000956_040 |
0.1033603524 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 15 |
Hb_000421_110 |
0.1048394536 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004526_010 |
0.1068711041 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |
| 17 |
Hb_003687_120 |
0.107185169 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 18 |
Hb_010775_010 |
0.1073586248 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004319_050 |
0.107754853 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 20 |
Hb_001369_030 |
0.109056017 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |