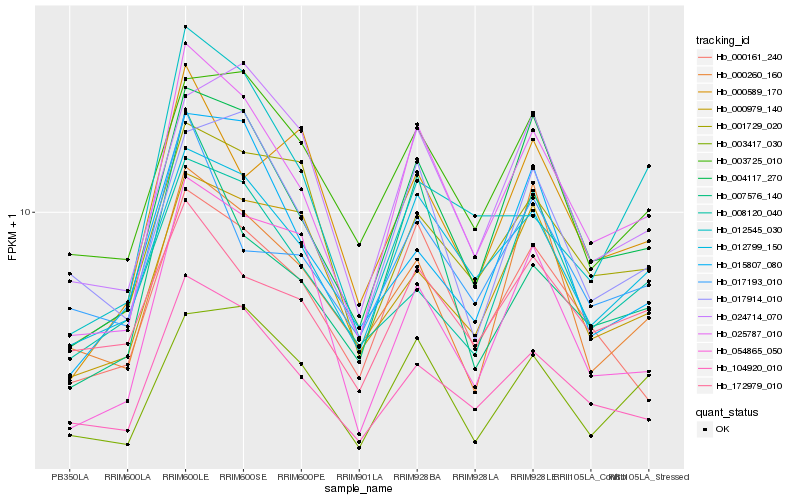
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025787_010 |
0.0 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 2 |
Hb_004117_270 |
0.0797502125 |
- |
- |
- |
| 3 |
Hb_104920_010 |
0.1002976909 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 4 |
Hb_012799_150 |
0.1035531978 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methyl-CpG-binding domain-containing protein 13 [Jatropha curcas] |
| 5 |
Hb_003417_030 |
0.1230335678 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000161_240 |
0.1238458712 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_054865_050 |
0.1325383831 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 8 |
Hb_001729_020 |
0.134471278 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_017914_010 |
0.1345920185 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 10 |
Hb_007576_140 |
0.1365835007 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 11 |
Hb_003725_010 |
0.1369998096 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 12 |
Hb_024714_070 |
0.1377729653 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 13 |
Hb_015807_080 |
0.1391519519 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 14 |
Hb_000260_160 |
0.1448776229 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000979_140 |
0.1458082667 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 16 |
Hb_172979_010 |
0.1478203993 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 17 |
Hb_000589_170 |
0.148013585 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_008120_040 |
0.1487984406 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 19 |
Hb_017193_010 |
0.1500628704 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_012545_030 |
0.1504230147 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |