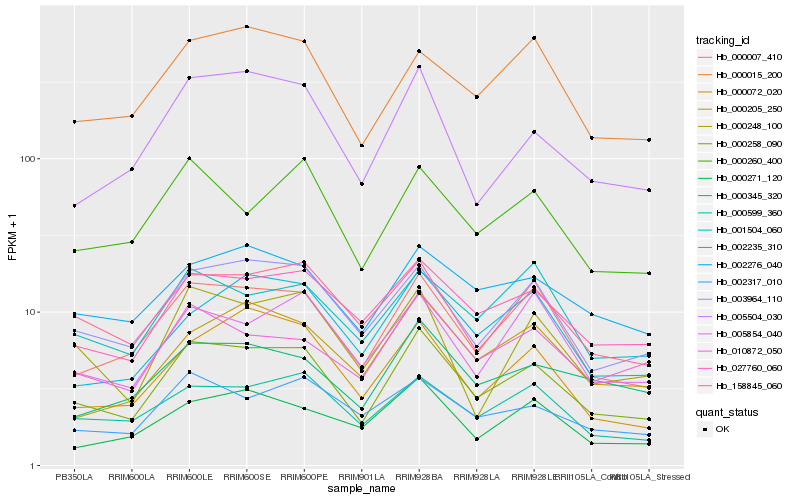
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_090 |
0.0 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000007_410 |
0.091153834 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 3 |
Hb_003964_110 |
0.0922400614 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 4 |
Hb_027760_060 |
0.0977764808 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 5 |
Hb_158845_060 |
0.1011333054 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000599_360 |
0.1054145352 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 7 |
Hb_005504_030 |
0.1085312202 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 8 |
Hb_002276_040 |
0.1117685854 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 9 |
Hb_000015_200 |
0.1159740326 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 10 |
Hb_002317_010 |
0.1166332461 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_000345_320 |
0.1169459075 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein ALWAYS EARLY 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_010872_050 |
0.1171553754 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 13 |
Hb_000260_400 |
0.1177685132 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 14 |
Hb_000072_020 |
0.1191174099 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000248_100 |
0.119920466 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 16 |
Hb_005854_040 |
0.1206072251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002235_310 |
0.1213289364 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 18 |
Hb_000205_250 |
0.1218384744 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 19 |
Hb_000271_120 |
0.122514905 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 20 |
Hb_001504_060 |
0.1226315593 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |