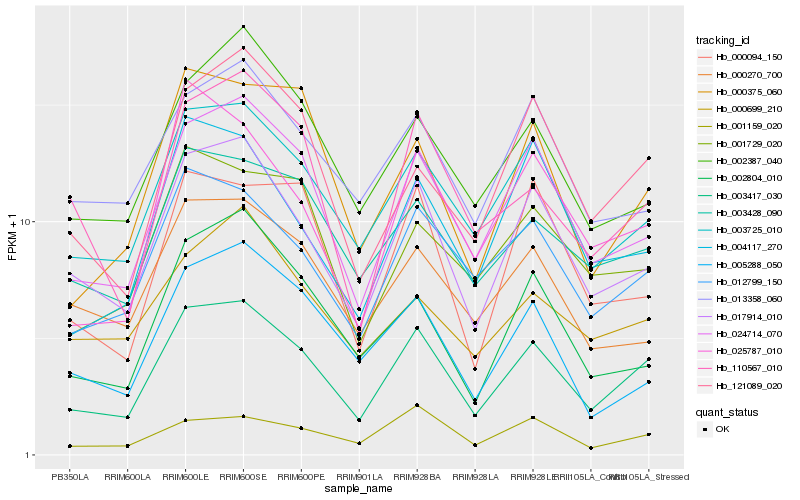
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003417_030 |
0.0 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_017914_010 |
0.0814295038 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 3 |
Hb_024714_070 |
0.0883311013 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 4 |
Hb_003725_010 |
0.0942801961 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 5 |
Hb_012799_150 |
0.1029554328 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methyl-CpG-binding domain-containing protein 13 [Jatropha curcas] |
| 6 |
Hb_003428_090 |
0.1172371946 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 7 |
Hb_121089_020 |
0.1215857258 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 8 |
Hb_001159_020 |
0.1229840712 |
- |
- |
- |
| 9 |
Hb_025787_010 |
0.1230335678 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 10 |
Hb_000375_060 |
0.1240793488 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 11 |
Hb_004117_270 |
0.125678916 |
- |
- |
- |
| 12 |
Hb_110567_010 |
0.1269015133 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_002387_040 |
0.1297259246 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 14 |
Hb_001729_020 |
0.1307907799 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_013358_060 |
0.1316274935 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 16 |
Hb_000094_150 |
0.1339778198 |
- |
- |
- |
| 17 |
Hb_000270_700 |
0.1355111402 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 18 |
Hb_005288_050 |
0.1360612539 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 19 |
Hb_002804_010 |
0.1360888554 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 20 |
Hb_000699_210 |
0.1370800565 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |