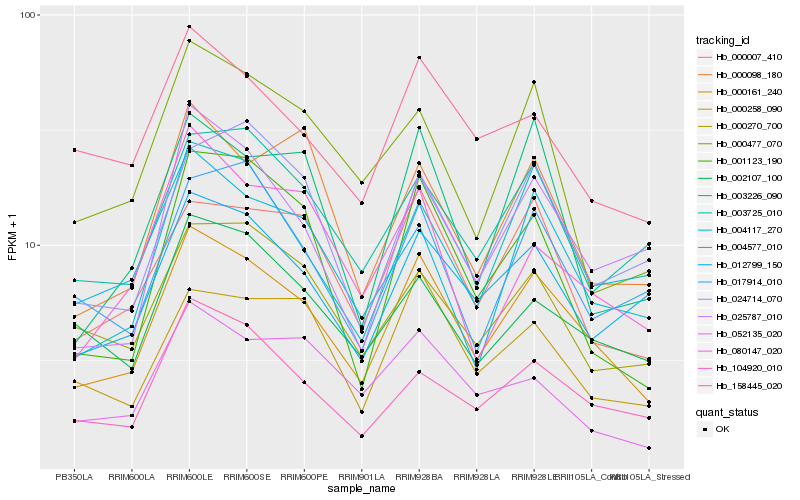
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000161_240 |
0.0 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000007_410 |
0.1118574267 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 3 |
Hb_104920_010 |
0.1121682039 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 4 |
Hb_012799_150 |
0.1140881805 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methyl-CpG-binding domain-containing protein 13 [Jatropha curcas] |
| 5 |
Hb_004117_270 |
0.1176829064 |
- |
- |
- |
| 6 |
Hb_158445_020 |
0.1224067451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000258_090 |
0.123340945 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 8 |
Hb_025787_010 |
0.1238458712 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 9 |
Hb_004577_010 |
0.1246439294 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 10 |
Hb_003725_010 |
0.1247480136 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 11 |
Hb_000270_700 |
0.1250130789 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 12 |
Hb_002107_100 |
0.1260375948 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 13 |
Hb_001123_190 |
0.1305543503 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 14 |
Hb_017914_010 |
0.1320869654 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 15 |
Hb_000098_180 |
0.1323341184 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003226_090 |
0.1327675135 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 17 |
Hb_024714_070 |
0.1334895878 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 18 |
Hb_000477_070 |
0.1337019661 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 19 |
Hb_080147_020 |
0.1353394108 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 20 |
Hb_052135_020 |
0.1354269721 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |