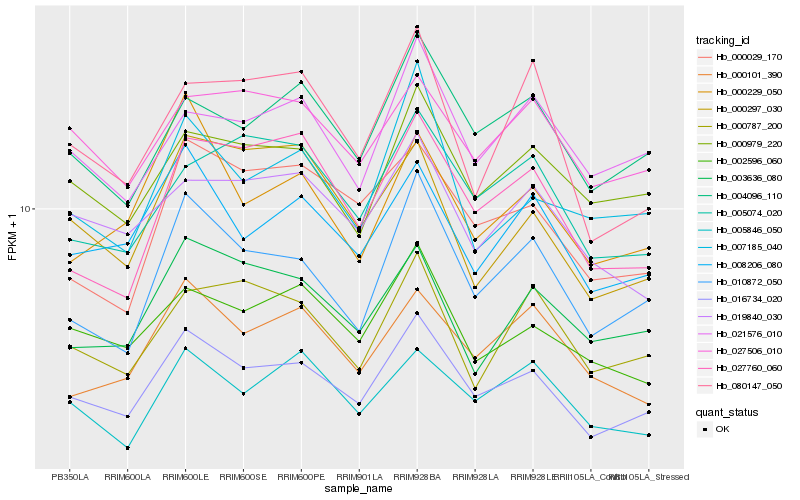
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016734_020 |
0.0 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_010872_050 |
0.0695738189 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 3 |
Hb_008206_080 |
0.0845544578 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 17 [Jatropha curcas] |
| 4 |
Hb_004096_110 |
0.0874335165 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 5 |
Hb_002596_060 |
0.0884401672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000979_220 |
0.092196776 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 7 |
Hb_027760_060 |
0.0933348464 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 8 |
Hb_000029_170 |
0.0943476034 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 89-like [Jatropha curcas] |
| 9 |
Hb_080147_050 |
0.0981615941 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 10 |
Hb_007185_040 |
0.0984512339 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 11 |
Hb_000229_050 |
0.0985390344 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 12 |
Hb_005074_020 |
0.0988795603 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003636_080 |
0.0990785144 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 14 |
Hb_019840_030 |
0.0993850662 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000101_390 |
0.09960424 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 16 |
Hb_021576_010 |
0.1013440445 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 17 |
Hb_000297_030 |
0.1021469054 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000787_200 |
0.1024173901 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_027506_010 |
0.102418182 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 20 |
Hb_005846_050 |
0.1028407485 |
- |
- |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |