| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
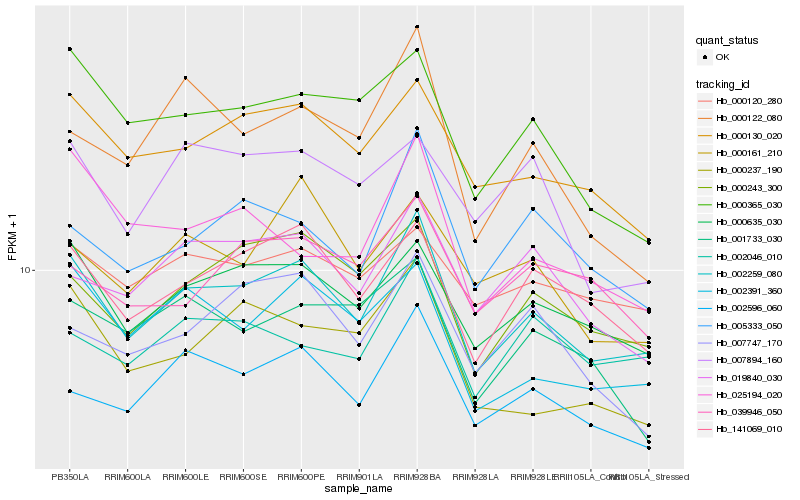
Hb_002259_080 |
0.0 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 2 |
Hb_000635_030 |
0.0718902069 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 3 |
Hb_141069_010 |
0.0786137951 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000130_020 |
0.0814143602 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 5 |
Hb_002596_060 |
0.0833365136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000243_300 |
0.0833676668 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_002391_360 |
0.0911382912 |
- |
- |
hypothetical protein CICLE_v10026208mg [Citrus clementina] |
| 8 |
Hb_005333_050 |
0.0945737567 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000365_030 |
0.0962668195 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 10 |
Hb_019840_030 |
0.0962998928 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000120_280 |
0.1005017849 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 12 |
Hb_002046_010 |
0.1020568739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000237_190 |
0.1035522901 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000161_210 |
0.1045462572 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 15 |
Hb_007747_170 |
0.1084978211 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 16 |
Hb_025194_020 |
0.1087548824 |
- |
- |
PREDICTED: uncharacterized protein C1F8.04c-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001733_030 |
0.108786791 |
- |
- |
PREDICTED: uncharacterized protein LOC100261386 isoform X2 [Vitis vinifera] |
| 18 |
Hb_000122_080 |
0.1093298476 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 19 |
Hb_039946_050 |
0.1096564219 |
- |
- |
catalytic, putative [Ricinus communis] |
| 20 |
Hb_007894_160 |
0.1099970397 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |