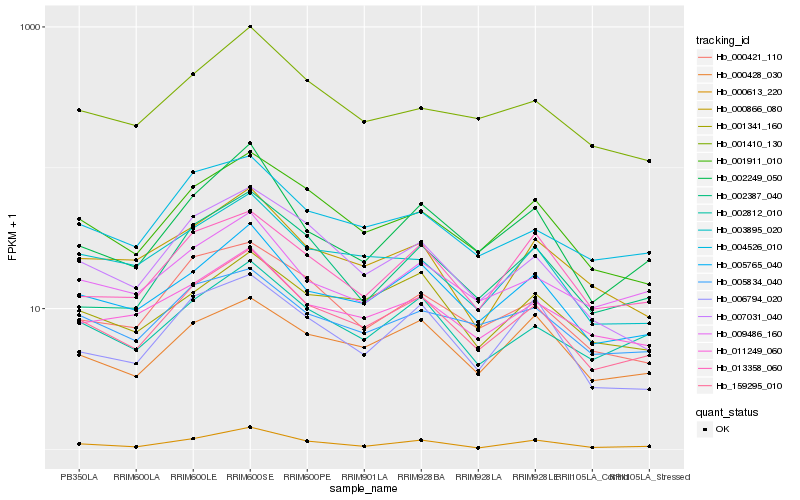
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_159295_010 |
0.0 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 2 |
Hb_005765_040 |
0.0554318762 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 3 |
Hb_001911_010 |
0.0655136952 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 4 |
Hb_002812_010 |
0.0791637302 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002249_050 |
0.0812666215 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 6 |
Hb_004526_010 |
0.0831181356 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |
| 7 |
Hb_011249_060 |
0.0849526401 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 8 |
Hb_003895_020 |
0.0882485186 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000421_110 |
0.0883854315 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001410_130 |
0.0887707515 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 11 |
Hb_007031_040 |
0.0894178574 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 12 |
Hb_001341_160 |
0.0914156202 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000428_030 |
0.0925996022 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 14 |
Hb_009486_160 |
0.0933207559 |
- |
- |
PREDICTED: metacaspase-1 [Jatropha curcas] |
| 15 |
Hb_000866_080 |
0.0947514226 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006794_020 |
0.0950018145 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 17 |
Hb_005834_040 |
0.0953911283 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 18 |
Hb_002387_040 |
0.0963069097 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 19 |
Hb_000613_220 |
0.0973528758 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 20 |
Hb_013358_060 |
0.0974028481 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |