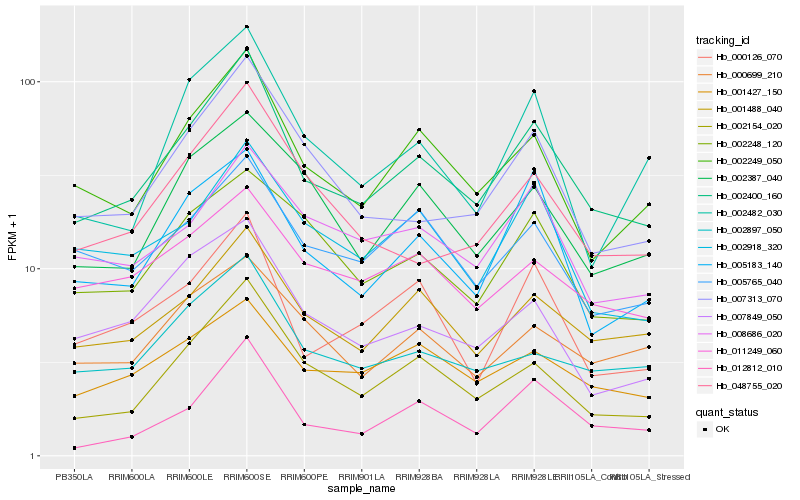
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_160 |
0.0 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 2 |
Hb_002249_050 |
0.0915933725 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 3 |
Hb_002154_020 |
0.0958120468 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_001488_040 |
0.0987912258 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 5 |
Hb_002897_050 |
0.1103735966 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 6 |
Hb_007313_070 |
0.1114162843 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 7 |
Hb_000126_070 |
0.1133900552 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Gossypium arboreum] |
| 8 |
Hb_005183_140 |
0.1153237663 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 9 |
Hb_001427_150 |
0.1158667881 |
- |
- |
PREDICTED: uncharacterized protein LOC105637790 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002918_320 |
0.1194497651 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 11 |
Hb_048755_020 |
0.1203921034 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 12 |
Hb_012812_010 |
0.1212221865 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 13 |
Hb_007849_050 |
0.1214738475 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 14 |
Hb_008686_020 |
0.1214761226 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002387_040 |
0.1231339629 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 16 |
Hb_002248_120 |
0.1247048116 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 17 |
Hb_000699_210 |
0.124747811 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 18 |
Hb_005765_040 |
0.1248306453 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 19 |
Hb_011249_060 |
0.1264609095 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 20 |
Hb_002482_030 |
0.1266287134 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |