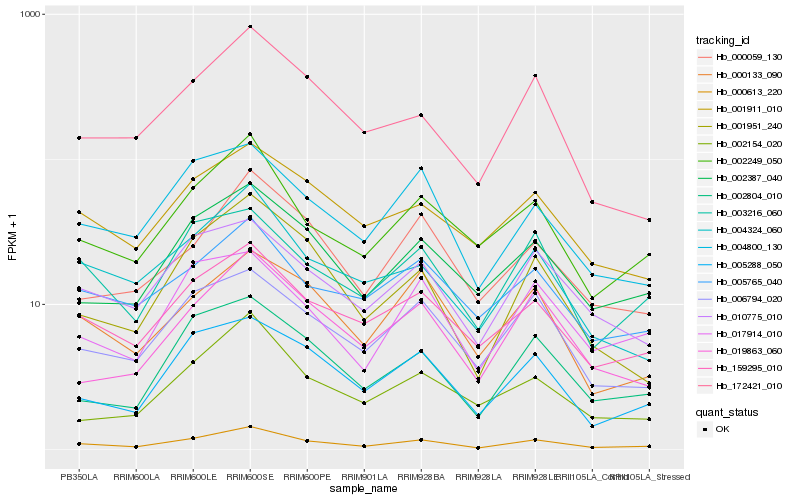
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000613_220 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 2 |
Hb_159295_010 |
0.0973528758 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 3 |
Hb_002249_050 |
0.0974731022 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 4 |
Hb_002387_040 |
0.1047072844 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 5 |
Hb_005288_050 |
0.1063256383 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 6 |
Hb_002804_010 |
0.1091000882 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 7 |
Hb_006794_020 |
0.1120569018 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 8 |
Hb_010775_010 |
0.1130497051 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003216_060 |
0.1145227981 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 10 |
Hb_001951_240 |
0.1145572322 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 11 |
Hb_004324_060 |
0.1151106486 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 12 |
Hb_004800_130 |
0.1159905453 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 13 |
Hb_019863_060 |
0.1200204664 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 14 |
Hb_017914_010 |
0.1200771217 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 15 |
Hb_000133_090 |
0.1214982915 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 16 |
Hb_172421_010 |
0.1216437798 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 17 |
Hb_002154_020 |
0.1220637153 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_005765_040 |
0.1223191144 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 19 |
Hb_000059_130 |
0.1229060835 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 20 |
Hb_001911_010 |
0.1233742894 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |