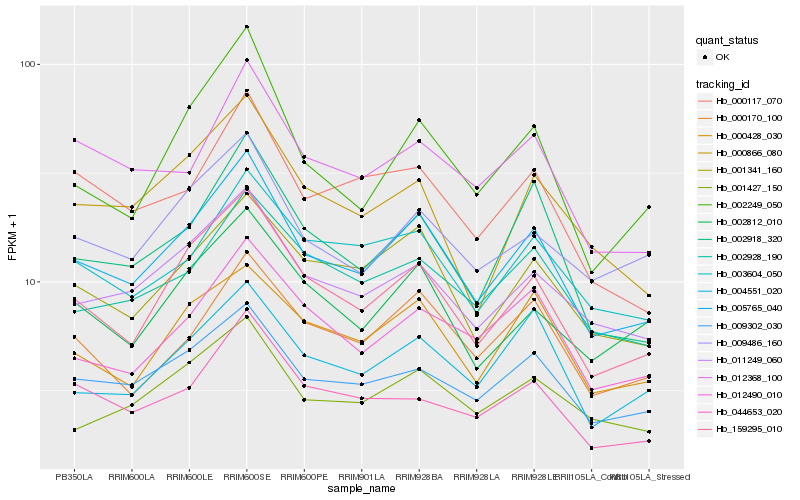
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005765_040 |
0.0 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 2 |
Hb_159295_010 |
0.0554318762 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 3 |
Hb_011249_060 |
0.0736202667 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 4 |
Hb_001341_160 |
0.0761501671 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_009302_030 |
0.0766787951 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 6 |
Hb_002918_320 |
0.0796386987 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003604_050 |
0.0864403569 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000170_100 |
0.0874625411 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 9 |
Hb_000428_030 |
0.0875892287 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 10 |
Hb_002928_190 |
0.0877884377 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 11 |
Hb_002249_050 |
0.0878524444 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 12 |
Hb_001427_150 |
0.0882583286 |
- |
- |
PREDICTED: uncharacterized protein LOC105637790 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009486_160 |
0.0887238341 |
- |
- |
PREDICTED: metacaspase-1 [Jatropha curcas] |
| 14 |
Hb_044653_020 |
0.0895067995 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 15 |
Hb_002812_010 |
0.0895363981 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_012368_100 |
0.0899037983 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 17 |
Hb_000117_070 |
0.0899606045 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 18 |
Hb_012490_010 |
0.0907413034 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000866_080 |
0.0909560718 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004551_020 |
0.0912509719 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |