| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
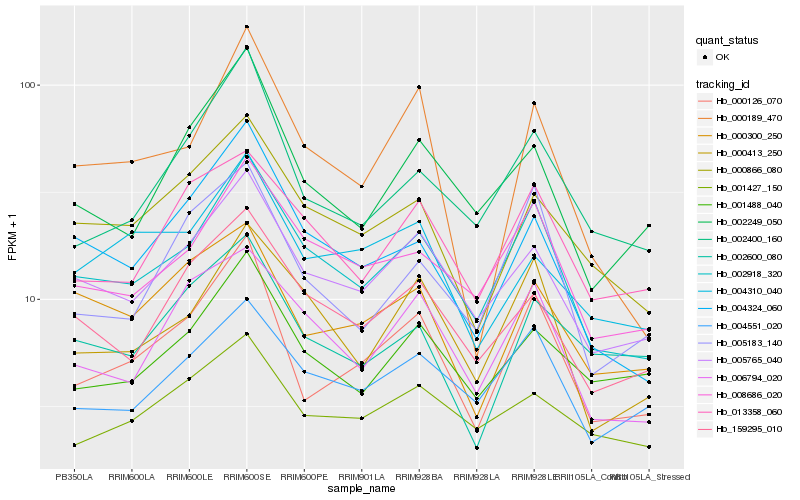
Hb_000126_070 |
0.0 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Gossypium arboreum] |
| 2 |
Hb_002400_160 |
0.1133900552 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 3 |
Hb_002918_320 |
0.1179250148 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005765_040 |
0.1266949541 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 5 |
Hb_005183_140 |
0.129749529 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 6 |
Hb_002249_050 |
0.1320225838 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 7 |
Hb_000300_250 |
0.1339923783 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000866_080 |
0.1406873949 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000189_470 |
0.1424376514 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 10 |
Hb_001488_040 |
0.1438583659 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 11 |
Hb_004551_020 |
0.1440996744 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 12 |
Hb_001427_150 |
0.1457913008 |
- |
- |
PREDICTED: uncharacterized protein LOC105637790 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004324_060 |
0.1458940047 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 14 |
Hb_008686_020 |
0.1469695723 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004310_040 |
0.1479743073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006794_020 |
0.1487415595 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 17 |
Hb_002600_080 |
0.1500386469 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000413_250 |
0.1515818063 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 19 |
Hb_013358_060 |
0.1517827916 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 20 |
Hb_159295_010 |
0.1518182163 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |