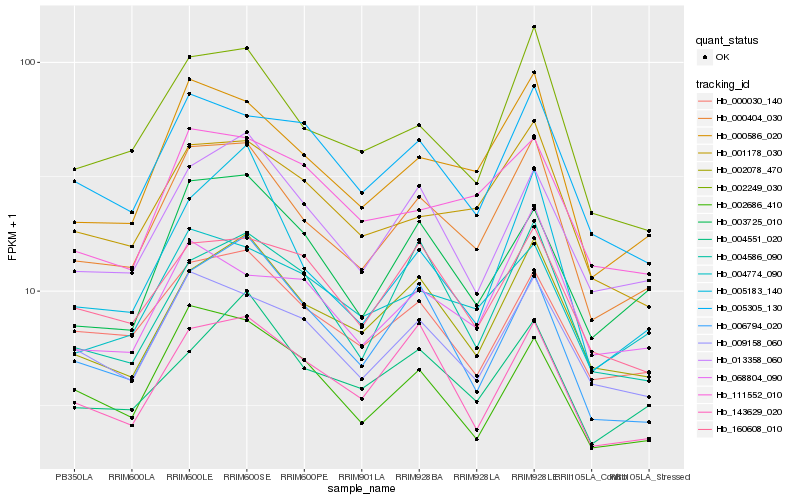
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000404_030 |
0.0 |
- |
- |
PREDICTED: protein NAP1 [Jatropha curcas] |
| 2 |
Hb_000586_020 |
0.0559822674 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein F383_02497 [Gossypium arboreum] |
| 3 |
Hb_002249_030 |
0.0692966805 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002078_470 |
0.0789782312 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001178_030 |
0.0859036109 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 6 |
Hb_009158_060 |
0.0864969565 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 7 |
Hb_013358_060 |
0.0892424384 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 8 |
Hb_000030_140 |
0.0907577827 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004774_090 |
0.0930612438 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 10 |
Hb_003725_010 |
0.0948576081 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 11 |
Hb_005305_130 |
0.0964024274 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 12 |
Hb_005183_140 |
0.0973778091 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 13 |
Hb_002686_410 |
0.0975591074 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_143629_020 |
0.0983347671 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 15 |
Hb_004551_020 |
0.1019147596 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 16 |
Hb_004586_090 |
0.103505445 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006794_020 |
0.1035731003 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 18 |
Hb_111552_010 |
0.1039792012 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 19 |
Hb_160608_010 |
0.1044648739 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 20 |
Hb_068804_090 |
0.1047339116 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |