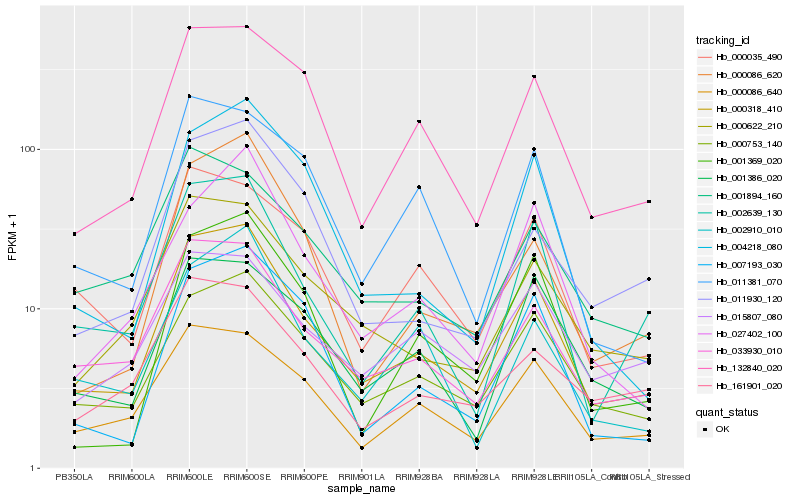
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_410 |
0.0 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 2 |
Hb_000753_140 |
0.0943971409 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 3 |
Hb_033930_010 |
0.0947957082 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 4 |
Hb_132840_020 |
0.1144966465 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 5 |
Hb_000622_210 |
0.1151445845 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_002639_130 |
0.1159471159 |
- |
- |
hypothetical protein JCGZ_08129 [Jatropha curcas] |
| 7 |
Hb_000086_640 |
0.1160385125 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 8 |
Hb_000035_490 |
0.1306550142 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 9 |
Hb_001386_020 |
0.1314674506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007193_030 |
0.1331069501 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000086_620 |
0.1368080226 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001369_020 |
0.1381092907 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 13 |
Hb_015807_080 |
0.1399843555 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 14 |
Hb_001894_160 |
0.1408632676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_161901_020 |
0.1413889778 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 16 |
Hb_027402_100 |
0.1414102897 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 17 |
Hb_011381_070 |
0.1419680062 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 18 |
Hb_011930_120 |
0.1427078662 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002910_010 |
0.1437287263 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 20 |
Hb_004218_080 |
0.1453000751 |
- |
- |
PREDICTED: uncharacterized protein LOC105631207 [Jatropha curcas] |