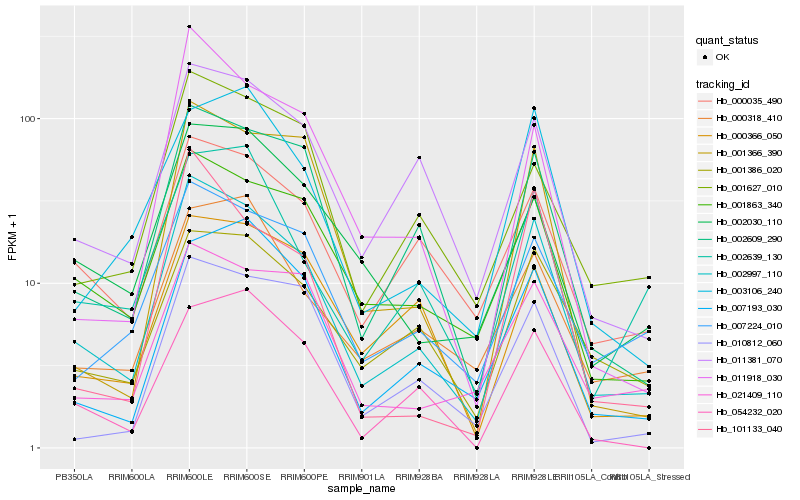
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002997_110 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002609_290 |
0.115671889 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 3 |
Hb_001863_340 |
0.1306337305 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 4 |
Hb_011381_070 |
0.1387924982 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 5 |
Hb_007224_010 |
0.1434528092 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 6 |
Hb_001386_020 |
0.1446995253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000035_490 |
0.1452806779 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 8 |
Hb_021409_110 |
0.1481186703 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_001627_010 |
0.1538532093 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 10 |
Hb_007193_030 |
0.1553082249 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001366_390 |
0.1576252348 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 12 |
Hb_010812_060 |
0.158157594 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 13 |
Hb_011918_030 |
0.1586087751 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 14 |
Hb_003106_240 |
0.1600884257 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 15 |
Hb_000318_410 |
0.1611394979 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 16 |
Hb_101133_040 |
0.1614397979 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_002639_130 |
0.1632835168 |
- |
- |
hypothetical protein JCGZ_08129 [Jatropha curcas] |
| 18 |
Hb_002030_110 |
0.163294409 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 19 |
Hb_000366_050 |
0.1636126033 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_054232_020 |
0.1641649562 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |